Publication Information
Frequency: Continuous
Format: PDF and HTML
Versions: Online (Open Access)
Year first Published: 2019
Language: English
| Journal Menu |
| Editorial Board |
| Reviewer Board |
| Articles |
| Open Access |
| Special Issue Proposals |
| Guidelines for Authors |
| Guidelines for Editors |
| Guidelines for Reviewers |
| Membership |
| Fee and Guidelines |
 |
The RhMYB24/bHLH71 complex coordinates flower opening by mediating auxin and ethylene crosstalk in Rosa hybrida
Nisar Hussain1,2*, Adeel Shahid3, Yangchao Jia1, Hafiz Ghulam Nabi4, Muhammad Ahmad5
1Beijing Key Laboratory of Development and Quality Control of Ornamental Crops, Department of Ornamental Horticulture, College of Horticulture, China Agricultural University, Beijing, 100193, China.
2Department of Production Technology, 24Hua Kunming Modern Agricultural Technology Co., Ltd, Kunming, China.
3Department of Agriculture, Government College University Lahore, 54000, Lahore, Pakistan.
4State Key Laboratory of Agrobiotechnology/Beijing Key Laboratory of Crop Genetic Improvement, College of Agronomy and Biotechnology, China Agricultural University, Beijing 100193, China.
5Department of Vegetable Science, College of Horticulture, China Agricultural University, Beijing 100193, China.
Received Date: September 30; Accepted Date: October 14, 2024; Published Date: November 09, 2024;
*Corresponding author: Nisar Hussain, Beijing Key Laboratory of Development and Quality Control of Ornamental Crops, Department of Ornamental Horticulture, College of Horticulture, China Agricultural University, Beijing, 100193, China. Email: nisar.384@hotmail.com; nisarhussain@cau.edu.cn
Citation: Hussain N, Shahid A, Jia Y, Nabi HG, Ahmad M; (2024) The RhMYB24/bHLH71 complex coordinates flower opening by mediating auxin and ethylene crosstalk in Rosa hybrida. Adv Agri Horti and Ento: AAHE-213.
DOI: 10.37722/AAHAE.2024401
Abstract
Rose is a leading cut flower in production but owing to improper flower opening, it is facing heavy losses in postharvest handling. This study aims to explore the regulatory networks of transcription factors and phytohormones involved in flower opening and their role in petal cell expansion. Here, we isolated a highly expressed transcription factor RhMYB24, whose expressions are induced by auxin and gibberellin, and inhibited by ethylene. The silencing of RhMYB24 downregulates cell expansion-related genes and delayed flower opening. We found that RhMYB24 interacts with the RhbHLH71 and controls flower openings in a complex. We uncovered that RhbHLH71 also has high expression during the early stages of flower opening, and found its expression was significantly lower in TRV-RhMYB24 plants compared to TRV plants. Further, silencing of RhbHLH71 resulted in smaller petal size and delayed flower opening. We observed that TRV-RhbHLH71 had lower expression of RhMYB24 and cell expansion-related genes. Furthermore, we exhibited that RhMYB24 and RhbHLH71 controlled genes related to auxin, GAs, and ethylene-regulated genes. Our results indicate that RhMYB24 and RhbHLH71 form a complex and coordinate flower opening through cell expansion genes by mediating the crosstalk between auxin and ethylene signaling.
Keywords: Rosa hybrida, flower opening, cell expansion, bHLH71, MYB24, auxin, ethylene
Background
Flower opening is a complex biological process that leads to pollination and determines the market value of cut flowers. Rose, is one of the leading cut flowers supplied by long-distance transportation, which disturbs proper flower opening and diminishes the ornamental period. Flower opening is mainly attributable to cell expansion (Yamada et al., 2009) via the actions of phytohormones, including auxin, gibberellic acid (GAs), and ethylene (Sun et al., 2021). Auxin is an essential phytohormone in cell division and expansion (Li et al., 2016). Exogenous application of auxin or its inhibitor can lead to changes in flower opening. The Auxin (IAA and NAA) applications in the iris encouraged elongation and flower opening while auxin inhibitor 2,3,5-triodobenzoic acid (TIBA) and α-(p-chlorophenoxy)-isobutyric acid; (PCIB) repressed cell elongation and flower opening (van Doorn et al., 2013). In waterlily, auxin transcriptome profiles during flower opening go high, while genes related to cell wall strength are downstream of auxin events gone down resulting in flower opening (Ke et al., 2018). The auxin-regulated RhARF2 is a critical player that controls flower opening by governing RhMYB6 expression and mediating the crosstalk between auxin and ethylene signaling (Chen et al., 2023). Ethylene represents a critical function at several stages of growth and development in plants. Ethylene production was measured in rose, and found that short-lived varieties reached the ethylene peak earlier than long-lived varieties (Xue et al., 2008). Applying ethylene to rose results in repressed petal cell expansion by directly inhibiting the aquaporin proteins, while ethylene inhibitor 1-MCP application resulted in more curved petals with larger cell size (Ma et al., 2008). GA promotes the elongation of petals and stamens through repressing DELLA proteins in Arabidopsis (Cheng et al., 2004). The antagonism between ethylene and GA during rose petal expansion is mediated through regulating GA levels and DELLA protein stability (Chen et al., 2020; Luo et al., 2013). Due to the importance of phytohormones in flower opening, it is critical to determine the regulatory mechanisms governing the interaction between different phytohormones.
A few proteins that regulate floral organ size have been identified. There are numerous MYB TFs participate in flower development (Dubos et al., 2010). Such as, AtMYB103 regulates pollen and tapetal development in addition to trichrome formation in Arabidopsis (Higginson et al., 2003). AtMYB21 ectopic expression in Arabidopsis produced smaller plants and had minor leaves (Shin et al., 2002). Mutation of AtMYB32 produces partially male-sterile flowers, probably due to disruption of the phenylpropanoid pathway (Preston et al., 2004). The MYB-related transcription factor MIXTA controls the development of conical epidermal cells in the petals of Antirrhinum majus (Baumann et al., 2007). AtMYB24 has a high expression level in flowers and controls anther development. The overexpression of AtMYB24 in Arabidopsis results in abnormal pollen grains and anthers (Yang et al., 2007). An R2R3 MYB TF, Carbon Starved Anther (CSA), plays a significant part in modulating the sugar compartment and promoting pollen grain development (Zhang et al., 2010, Zhu et al., 2015). Several bHLH proteins are influenced by auxin and play an important role in flower growth and development (Groszmann et al., 2008). The petal-specific helix-loop-helix (bHLH) transcription factor BIGPETALp (BPEp) modulates petal expansion by specifically restricting cell expansion (Szécsi et al., 2006). The SPATULA is a bHLH protein that controls the growth of various tissues of plants and is involved in leaf, and petal expansion (Groszmann et al., 2010). However, the role of MYB and bHLH family proteins and regulations by phytohormone in flower opening largely remains unknown.
In this study, we found that a RhMYB24 transcription factor interacts with RhbHLH71 and orders the process of flower opening. The RhMYB24/bHLH71 complex regulates the genes related to cell expansion and mediates the crosstalk between auxin, GAs, and ethylene signaling.
Materials and Methods
Plant materials and treatments
Micropropagated Rosa hybrida ‘Samantha’ plantlets were grown on Murashige and Skoog (MS) medium with 3% (w/v) sucrose and 0.7% (w/v) agar as basal medium, supplemented with 1.0 mg·L−1 6-benzyl amino purine (6-BA), 0.05 mg·L−1 α- naphthalene acetic acid (NAA), and 3 mg·L−1 gibberellin acid (GA 3). Plantlets were incubated at (22 ±1) °C under a 16 h light and 8 h dark photoperiod (Zhang et al., 2019). Four-week-old plantlets were shifted to 1/2 MS medium supplemented with 0.1 mg ·L−1 NAA for rooting. After 3 weeks, the rooted seedlings were transferred to pots with vermiculite and peat moss in a 1:1 ratio under a 16 h light/8 h dark photoperiod at (22 ±1) °C with a relative humidity of 60%.
The flower opening stages for expression pattern analysis were defined as previously described (Ma et al., 2005). Petal samples at different stages (stages 1–5) were picked from the same middle whorl of the flowers (Wu et al., 2017). Cut rose flowers at stage 2 were used for hormone treatments. For hormone treatments, cut rose flowers were placed in vases containing 80 μmol·L−1 GA 3, 100 μmol·L−1 6-BA, and 100 μmol·L−1 NAA for 24 h. Mock samples were placed in 0.1% dimethyl sulfoxide. For ethylene treatment, cut rose flowers in vases were exposed to 10 μL·L−1 ethylene in a sealed 96 L glass box (0.4 m ×0.4 m ×0.6 m) for 24 h. Control flowers were sealed with air. Three biological replicates were collected for each sample.
Cloning and sequence analysis
The RhMYB24 Open Reading Frame (ORF) was amplified from R. hybrida ‘Samantha’ petal. Primers (Table 1) were designed by Primer Premier 5.0 software based on the predicted RchiOBHm_Chr2g0172331 sequence from the R. chinensis ‘Old Blush’ genome (Raymond et al., 2018). The sequences of its putative proteins in other plants were obtained from the NCBI database. Sequence alignment of RhMYB24 with its homologs from different plants was performed using ClustalW (https://www. genome.jp/tools-bin/clustalw) with default parameters. A phylogenetic tree was constructed with MEGA7.0 software using the neighbor-joining method by bootstrap analysis in 1,000 replicates.
RNA extraction and quantitative RT-PCR analysis
Total RNA was extracted from rose petals using the hot borate method as previously described (Chen et al., 2023). According to the manufacturer’s instructions, cDNA was synthesized from 1 μg total RNA using M-MLV reverse transcriptase (Promega). Quantitative RT-PCR (qRT-PCR) was performed in an Applied Biosystems StepOnePlus TM Real-Time PCR system using a KAPA SYBR FAST Universal qRT-PCR kit (Kapa Biosystems). The RhUBI2 gene was used as an internal control (Meng et al., 2013) and all reactions were performed in triplicate. The primers used for qRT-PCR are listed in Supplementary Table 1.
Virus-induced gene silencing
Virus-induced gene silencing (VIGS) was performed as previously described (Tian et al., 2014). A specific fragment from the ORF region and downstream region 406 bp in length of RhMYB24 containing EcoRI and BamHI restriction sites was used to construct the pTRV2-RhMYB24 vector. Agrobacterium tumefaciens strain GV3101 carrying the pTRV2-RhMYB24 vector was grown in Luria–Bertani (LB) medium supplemented with 50 μg·mL−1 kanamycin and 50 μg·mL−1 rifampicin, and then shaken in liquid medium at 28 °C at 200 r·min−1 overnight for 12–16 h. Agrobacterium cells were collected by centrifugation at 4,000 r·min−1 and re-suspended in infiltration buffer [10 mmol·L−1 2-(N-Morpholino) ethanesulfonic acid (MES), 200 mmol·L−1 acetosyringone, and 10 mmol·L−1 MgCl2, with pH 5.6] with a final OD 600 = 0.8–1.0. The pTRV1 and pTRV2-RhMYB24 were mixed at a ratio of 1:1 (v/v) (pTRV1 and pTRV2 were mixed separately as a control) and put in the dark at room temperature for 3–4 h. For inoculation, at least 100 rose seedlings were immersed in an infiltration buffer (0.4 L) and exposed to a vacuum of −25 kPa for 10 min. After the release of the vacuum, the plantlets were briefly washed with the same volume of deionized water and then incubated in the dark at 8 °C for 3 days. Then the plantlets were planted in pots and grown at (22 ±1) °C with a relative humidity of 60% and a 16 h light/8 h dark photoperiod for about 40 days. The phenotypes of flowers were monitored at various time points from stage 1 to stage 5.
For RhbHLH71 silencing, another specific fragment 251 bp in the length of RhbHLH71 was cloned and inserted in TRV2 between EcoRI and BamHI restriction sites and constructed the pTRV2-RhbHLH71 vector. The procedure was the same as in RhMYB24 silencing.
Microscopic observation and cell counting
Abaxial sub-epidermal (AbsE) cell photographs and counting were performed as previously described (Ma et al., 2008). Petal samples were taken as 5 mm diameter discs at 50% of the petal length from the top and fixed by formaldehyde-acetic acid (FAA) solution (3.7% formaldehyde, 5% glacial acetic acid, 50% ethanol, v/v). The number of AbsE cells was counted using ImageJ (National Institutes of Health, USA) per visual field.
Transactivation and Dual-Luciferase Reporter Assays
We examined the transactivation activity of the RhMYB24 protein with RhbHLH71 as described by Hellens (2005). we cloned the RhMYB24 protein into the pGreenII0800-LUC vector for determination expression of the luciferase reporter and inserted the ORFs RhbHLH71 into the pGreenII0029 62-SK vector to generate effector constructs under the control of the 35S promoter. Then transfer constructs into GV3101 (Agrobacterium strain) concealing the pSoup plasmid. After 3 d of infiltration, we harvested the N. benthamiana infiltrated leaves and sprayed them with 50 mg L21 D-luciferin (Promega). We captured images of LUC signals with a charge-coupled device camera (CHEMIPROHT 1300B/LND, Roper Scientific). For dual luciferase reporter assays, we co-infiltrated the above-described mixtures with 35Spro: REN constructs into N. benthamiana leaves. Three days later, we inspected LUC and REN activity with a GloMax luminometer and dual-luciferase reporter assay reagents of Promega.
Subcellular Localization of RhMYB24
To determine the subcellular localization of RhMYB24 in N. benthamiana leaf epidermal cells. We inserted the ORF of RhMYB24 in the pSuper1300 vector to form SUPERpro:RhMYB24-GFP, then introduced the vector into Agrobacterium strain GV3101. We infiltrated Agrobacterium cell suspensions into N. benthamiana leaves. After three days, we recorded the fluorescence signal on a laser confocal fluorescence microscopy (Olympus Fluo View FV1000). The GFP fluorescence signals were observed at an excitation wavelength of 488 nm and emission wavelength of 506-538 nm, and mCherry signals were detected using excitation with a 587-nm laser and emission with a 575- to675-nm band pass filter (Nelson et al., 2007).
Yeast two-hybrid assay
A cDNA library from the early stages of rose flower opening was used to screen out interacting proteins of RhMYB24 by using a yeast two-hybrid (Y2H) system. The coding sequence of RhMYB24 was inserted into the pGBKT7 (BD) vector as bait while the coding sequence of RhbHLH71 was introduced into the pGADT7 (AD) vector as prey. The bait and prey vectors were co-transformed into the yeast Gold strain. The transformed yeast was plated on SD/-Trp-Leu-His-Ade, Aureobasidin A (AbA), and X-gal plates.
Statistical analysis
The data were mean ±SD from at least three biological replicates for statistical analysis. Statistical analyses were performed using Student’s t-test with GraphPad Prism 8 (GraphPad Software Inc. San Diego, CA. USA). Statistically significant differences are indicated with P < 0.05 (∗) and P < 0.01 (∗∗).
Results
RhMYB24 was expressed during flower opening and responded to phytohormones.
During flower opening the petals expand rapidly to expose the anther and stigma for pollination. In ornamental, the flower’s opening speed is closely associated with their opening quality and ornamental period. We screened candidate genes to reveal the molecular mechanisms of flower opening and explored the key regulatory genes mainly focused on transcription factors (Chen et al., 2023). We found that a transcript RchiOBHm_Chr2g0172331 increased accompanied by rose flower opening (Fig. 1A). This gene was highly expressed in petals as compared to other floral organs (Fig. 1B), whose expressions were induced by auxin and gibberellin, and inhibited by ethylene (Fig. 1C).
Then, we isolated it from the petals of R. hybrida ‘Samantha’ whose ORF length was 618 bp, encoding a putative protein of 206 amino acids. Compared to its homologs in other species, the predicted protein of RchiOBHm_Chr2g0172331 contained a highly conserved R2R3-MYB domain that was homologous to AtMYB24 in Arabidopsis (Fig. 1D). Therefore, we named RchiOBHm_Chr2g0172331 as RhMYB24. Further, we found that it was localized in the nucleus and worked as a regulatory protein (Fig. 1E).
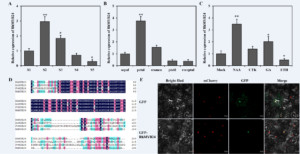
Figure 1: RhMYB24 was expressed during flower opening and responded to phytohormones.
(A) qRT-PCR analysis of rose transcript RchiOBHm_Chr2g0172331/RhMYB24 at different flower opening stages. (B) qRT-PCR analysis of RhMYB24 in different floral organs. (C) qRT-PCR analysis of RhMYB24 under different hormone treatments in petals. Flowers at stage 2 were treated with NAA, CTK, GA, and Ethylene (ETH). (D) Multiple sequence alignment of putative MYB24 protein in different species. Fv, Fragaria vesca; At, Arabidopsis thaliana; Md, Malus domestica. (E) Subcellular Localization of RhMYB24 in N. benthamiana leaf epidermal cells on a laser confocal fluorescence microscopy. Results are the means of three biological replicates from different plants with SE. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P < 0.1, **P < 0.01).
RhMYB24 controls flower opening.
To determine the functional role of RhMYB24 in the flower opening process, we did VIGS in Samantha rose. The silencing was based on the relative expression of RhMYB24 that was observed using qRT-PCR (Fig. 2A). We found that TRV-RhMYB24 plants had delayed flower opening (10 days) compared to TRV control plants (7 days) as shown in Figures 2B and 2D. Further, we found that the flower diameter is significantly reduced in TRV-RhMYB24 which starts from the 4th day and continues to the last day of opening (Figure 2C). These results indicate that RhMYB24 plays an important role in flower opening.

Figure 2: RhMYB24 controls flower opening
(A) Expression of RhMYB24 in the petals of TRV control plants and RhMYB24-silenced (TRV-RhMYB24) plants. (B) Statistics of longevity (number of days) in TRV control and RhMYB24-silenced plants during different flower opening periods. (C) Diameters of TRV control and RhMYB24-silenced flowers were measured daily until stage 5. (D) Phenotypes of TRV control and RhMYB24-silenced plants during flower opening were recorded daily. Images of flowers were photographed separately at a uniform scale. Results are the means of three biological replicates from different plants with SE. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P < 0.1, **P < 0.01).
RhMYB24 promoted petal cell expansion by regulating genes related to cell expansion.
Petal cell expansion promotes flower opening in roses (Yamada et al., 2009; Sun et al., 2021). We observed that RhMYB24 silenced plants have lower relative petal area than TRV control plants (Figure 3A, 3B). Further, with microscopy, we measured the cell area of petals and spotted that the average cell area was larger in TRV control plants while the cell numbers were less in the unit area of petals than in TRV-MYB24 plants (Figure 3C, 3D).
Furthermore, to determine the role of RhMYB24 in cell expansion at the molecular level we inspected the transcription level of different genes involved in cell expansion and cell wall integrity using qRT-PCR. We observed that compare to control the RhMYB24 silenced plants had relatively lower transcript level of RhPIP1;1, RhPIP2;1, RhCesA2, RhEXPA4, RhEXPA8, RhXTH6, RhTOR3, RhTOR4 genes (Figure 3E).
These results suggest that RhMYB24 affects the final size of petals by regulating petal cell expansion genes.
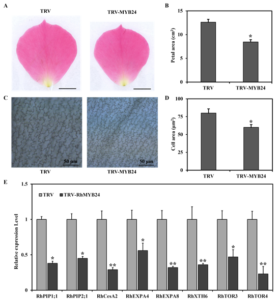
Figure 3: RhMYB24 promoted petal cell expansion by regulating genes related to cell expansion
(A) Petal of TRV control (left) and RhMYB24-silenced (right) rose plants. Images of RhMYB24-silenced and TRV control petals were photographed at stage 3. Scale bar represents 1 cm. (B) Petal sizes of TRV control and RhMYB24-silenced plants at a fully open stage. (C) Abaxial sub-epidermal (AbsE) cells in the middle regions of rose petals at stage 5. The scale bar represents 50 µm. (D) Measurement of TRV control and RhMYB24-silenced AbsE cell sizes at stage 5. (E) Expression of genes related to cell expansion in petals of TRV control and RhMYB24-silenced plants. Results are the means of three biological replicates from different plants with SE. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P < 0.05, **P < 0.01).
Screening of RhMYB24 interacting Y2H cDNA library from rose petals
To find proteins interacting with RhMYB24 and have a role in petal cell expansion, we screened a cDNA library from the early stage of flower-opening petals by a yeast two-hybrid (Y2H) system. We found that RhMYB24 interacted highly with proteins such as Beta-galactosidase 1 (BGAL1), Aspartate kinase 3 (AK3), Fructose-bisphosphate aldolase 6 (FBA6), basic helix-loop-helix proteins 71 (bHLH71), Pathogenesis-related thaumatin superfamily protein (PRT), Cellulose synthase-like A15 (CSLA15), Pectin lyase-like, Responsive to dehydration 21A (RD21A), ERF71-AP2-EREBP, and Thioredoxin f-type 1 (TRXF1) as shown in Table Responsive to dehydration 21A (RD21A), ERF71-AP2-EREBP, and Thioredoxin f-type 1 (TRXF1) as shown in Table 1. These proteins are active parts of different physiological pathways like the Calvin cycle, osmotic regulation, cell wall integrity, and photoperiodic regulation and play critical roles in plant growth. Among these proteins, bHLH71 is a regulatory protein localized in the nucleus (AT5G46690 arabidopsis.org). The ORF length of bHLH71 was 963 bp, encoding a putative protein of 321 amino acids, and its expression was highly changed in RhMYB24 silenced plants (Table 1, Figure 5A).
Then, to analyze the transcription regulatory activity of RhMYB24 with RhbHLH71 we did Y2H with Aureobasidin and X-gal, and dual-luciferase reporter assay in Nicotiana benthamiana and found that together they have strong transcription activity as compared to empty vectors (Figure 4A, 4B). Besides, RhMYB24 with RhbHLH71 showed strong LUC/REN activity (Figure 4C). These results indicate that RhMYB24 directly binds with RhbHLH71 and promotes its transcription.
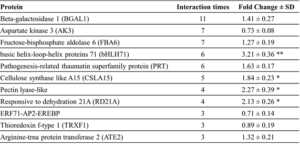
Table 1: RhMYB24 interaction with other proteins (Y2H)
To find proteins interacting with RhMYB24, a cDNA library from the early stage of flower-opening petals was screened by a yeast two-hybrid (Y2H) system. Here list of the most interacting proteins, with the interaction times, and their transcript level change in TRV-RhMYB24 plants. Results are the means of three biological replicates from different plants with SE. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P < 0.1, **P < 0.01).
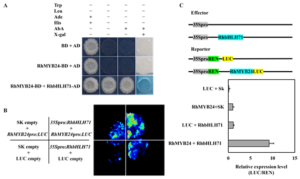
Figure 4: RhMYB24 interact with RhbHLH71
(A) RhMYB24 interacts with RhbHLH71 in the Yeast 2 Hybrid (Y2H) system. The Open reading frame (ORF) sequence of RhMYB24 was inserted into pGBKT7 (BD) as the bait, and the ORF sequence of RhbHLH71 was inserted into pGADT7 (AD) as the prey vector and platted on SD/-Trp-Leu-His-Ade + AbA + X-gal plates (B) Transactivation of the RhMYB24 protein with RhbHLH71. RhMYB24:LUC construct was co-infiltrated with 35S:RhbHLH71 protein or SK empty vector in N. benthamiana leaves. The representative image was shown 3 d after infiltration. (C) Dual-luciferase assays of RhMYB24 and RhbHLH71 proteins. Top, schematic representation of Effectors and Reporter. The bottom is a quantitative analysis of the transcriptional activation of the RhMYB24 with RhbHLH71 that were co-infiltrated into N. benthamiana leaves. The 35S promoter-derived Renilla luciferase (REN) gene was used as an internal control. Each data point represents mean ± SE (n ≥ 5). Different letters indicate significant differences (P < 0.05), as determined by one-way ANOVA.
RhbHLH71 controls flower opening
To explore the role of RhbHLH71 during flower opening, we done qRT-PCR and observed its transcription level in different floral parts and at different stages of flower opening. We found the RhbHLH71 transcript was highly expressed in petals compared to other floral parts (Figure 5B). The transcript was high at the early stages of flower opening and went down when the flower fully opened (Figure 5C). Additionally, to know about the pathway of how RhbHLH71 participates in flower opening we checked its response against different phytohormones. We found its transcripts induced by Auxin and highly repressed by ethylene (Figure 5D).
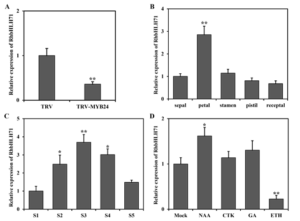
Figure 5: RhbHLH71 was expressed during flower opening and responded to phytohormones
(A) qRT-PCR analysis of RhbHLH71 in RhMYB24 silenced plants. (B) qRT-PCR analysis of RhbHLH71 in different floral organs. (C) qRT-PCR analysis of rose transcript RhbHLH71 at different flower opening stages. (D) qRT-PCR analysis of RhbHLH71 under different hormone treatments in petals. Flowers at stage 2 were treated with NAA, CTK, GA, and Ethylene (ETH). Results are the means of three biological replicates from different plants with SE. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P < 0.1, **P < 0.01).
To find the functional role in flower opening, we performed VIGS and found bHLH71 relative transcripts level significantly reduced in TRV-bHLH71 compared to TRV control (Figure 6A). The flower opening was delayed in silenced plants (10.44 days) compared to TRV control plants (7.42 days) as shown in Figures 6B and 6D. Further, we measured flower diameter daily and found that in TRV-bHLH71 plants flower diameter greatly reduced compared to TRV control plants (Figure 6C). These results showed that RhbHLH71 plays an important role in flower opening.

Figure 6: RhbHLH71 controls flower opening
(A) Expression of RhbHLH71 in the petals of TRV control plants and RhbHLH71-silenced (TRV-bHLH71) plants. (B) Statistics of longevity (number of days) in TRV control and RhbHLH71-silenced plants during different flower opening periods. (C) Diameters of TRV control and RhbHLH71-silenced flowers were measured daily. (D) Phenotypes of TRV control and RhbHLH71-silenced plants during flower opening were recorded daily. Images of flowers were photographed separately at a uniform scale. Results are the means of three biological replicates from different plants with SE. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P < 0.1, **P < 0.01).
RhbHLH71 controls flower opening by regulating petal cell expansion
To determine the role of RhbHLH71 in petal cell expansion we observed the petal area in TVR control and TRV- RhbHLH71 at stage 3. We found the petal area reduced in silenced petals compared to the control (Figure 7A, 7B). Further, microscopic petal observation showed that the AbsE cell area in RhbHLH71-silenced plants was significantly smaller at stage 3 (Figure 7C, 7D).
Furthermore, to determine the role of RhbHLH71 in cell expansion at the molecular level we inspected the transcription level of different genes involved in cell expansion and cell wall integrity using qRT-PCR. We observed that relative transcript level of RhPIP1;1, RhPIP2;1, RhCesA2, RhEXPA4, RhEXPA8, RhXTH6, RhTOR3, RhTOR4 genes were reduced in the RhbHLH71 silenced plants compared to TRV control (Figure 7E). These results suggest that RhbHLH71 affects the petal size by regulating petal cell expansion.
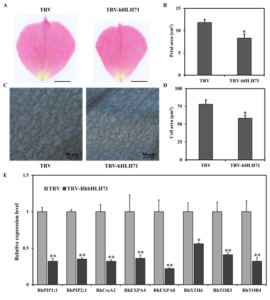
Figure 7: RhbHLH71 promoted petal cell expansion by regulating genes related to cell expansion.
(A) Petal of TRV control (left) and RhbHLH71-silenced (right) rose plants. Images of RhbHLH71-silenced and TRV control petals were photographed at stage 3. Scale bar represents 1 cm. (B) Petal sizes of TRV control and RhbHLH71-silenced plants at a fully open stage. (C) Abaxial sub-epidermal (AbsE) cells in the middle regions of rose petals at stage 5. The scale bar represents 50 µm. (D) Measurement of TRV control and RhbHLH71-silenced AbsE cell sizes at stage 5. (E) Expression of genes related to cell expansion in petals of TRV control and RhbHLH71-silenced plants. Results are the means of three biological replicates from different plants with SE. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P < 0.05, **P < 0.01).
RhMYB24 and RhbHLH71 mediate phytohormones crosstalk
As we saw from previous results RhMYB24 and RhbHLH71 were induced by auxin and repressed by ethylene and acted together in a coordinated way to control flower opening by regulating petal cell expansion. Further, to observe whether they mediate crosstalk of phytohormone we measured transcript levels of different phytohormone-related genes who participated in petal cell expansion. We found that silencing of one gene also lowers the transcript level of other genes (Figure 8A, 8B). The transcript level of auxin-related genes RhYUCCA6, RhIAA14, RhGH3, and Rh15A-like were lower in both RhMYB24 and RhbHLH71 silenced petals (Figure 8C, 8D). We observed that GAs biosynthesis gene RhGA20ox expression went down, while GAs biosynthesis inhibited gene RhGA2ox expression gone up in silenced plants (Figure 8A, 8B). We also found that an ethylene-regulated gene RhTIP1;1 promotes cell expansion its expression goes down, while the ethylene-regulated protein that suppresses cell expansion RhGAI1, expression goes up (Figure 8A, 8B). These results signify that RhMYB24 and RhbHLH71 form a complex and coordinate flower opening by mediating the crosstalk among phytohormones auxin, GAs, and ethylene-regulated genes, which participate in petal cell expansion.

Figure 8: RhMYB24 and RhbHLH71 mediate phytohormones crosstalk
(A) qRT-PCR analysis of RhbHLH71 in RhMYB24 silenced plants. (B) qRT-PCR analysis of RhMYB24 in RhbHLH71 silenced plants. (C-D) Transcript levels of different phytohormone-related genes that participated in petal cell expansion. Auxin-related genes RhYUCCA6, RhIAA14, RhGH3, and Rh15A-like. GAs biosynthesis gene RhGA20ox and GAs biosynthesis inhibited gene RhGA2ox and ethylene-regulated gene RhTIP1;1 and RhGAI1. Results are the means of three biological replicates from different plants with SE. Asterisks indicate statistically significant differences, as determined by Student’s t-test (*P < 0.1, **P < 0.01).
Discussion
Flower opening is a biological process that leads to pollination and determines the market value of cut flowers. Rose, is one of the leading cut flowers whose ornamental value mainly depends on the petals. Being supplied by long-distance transportation, flowers cannot open properly which diminishes the vase period. Flower opening is primarily attributable to cell expansion (Yamada et al., 2009) via the actions of phytohormones, including auxin, gibberellic acid, and ethylene (Sun et al., 2021).
During the petal expansion phase, genes associated with cell growth, such as genes encoding pectinesterases, extensins, and aquaporins, are upregulated (Bey et al., 2004; Pei et al., 2013). Auxin is essential for intrinsic developmental processes, including cell division, expansion, and differentiation (Strader and Zhao, 2016). During petal growth and expansion, auxin-related genes are upregulated to promote cell elongation by activating plasma membrane phosphorylation (Wang et al., 2017; Du et al., 2020), such as exogenous NAA application promotes cell elongation and flower opening in roses (Chen et al., 2023), but many details remain unknown. GAs are recognized as growth-promoting hormones in the cell elongation of plants (de Lucas et al., 2008). In rose, GAs through increasing the expression of RhGA20ox control petal cell expansion (Chen et al., 2020). It’s found that in roses, ethylene inhibits the cell expansion of petals during flower opening (Ma et al., 2008), and several transcription factors, such as RhGAI1 and NAC100, were intricate in ethylene-regulated petal cell expansion (Luo et al., 2013; Pei et al., 2013). Based on this finding, we identified the novel transcription factor RhMYB24, with abundant transcript levels during the early flower opening stage (Figure 1). The RhMYB24 silenced plants had delayed flower opening compared to the TRV control (Figure 2). The silenced plants have relatively small petals with reduced petal cell area (Figure 3). The results aligned with RhMYB6 which is induced by auxin and promotes petal cell expansion (Chen et al., 2023).
Cell expansion is a comprehensive biological process involved in the degradation and synthesis of cell walls, changes in cell turgor, and the reconstruction of the cytoskeleton (Cosgrove, 2005). Aquaporins are the major channel for water uptake across the plasma and intracellular membranes (Chaumont and Tyerman, 2014; Verdoucq et al., 2014). Earlier studies observed that two aquaporins, RhPIP1;1 and RhPIP2;1, were involved in petal cell expansion in rose (Ma et al., 2008; Chen et al., 2013). Expansins and xyloglucan endotransglycosylases/hydrolases (XTH) are well-known plant cell wall-loosening proteins (Rose et al., 2002). CESA genes are widely reported to participate in the process of cell wall synthesis (Mutwil et al., 2008). We observed that compared to the control the RhMYB24 silenced plants had relatively lower transcript levels of cell expansion-related genes such as RhPIP1;1, RhPIP2;1, RhCesA2, RhEXPA4, RhEXPA8, RhXTH6, RhTOR3, and RhTOR4 (Figure 3E). In earlier studies, we found that auxin and GAs promote petal cell expansion by regulating transcription factors MYB6, and NF-YC-9, which control petal cell expansion genes (Chen et al., 2020; Chen et al., 2023). These results suggest that RhMYB24 affects the final size of petals by regulating petal cell expansion.
The results of a cDNA library screening showed RhMYB24 interacted highly with proteins which are active parts of different physiological pathways like the Calvin cycle, osmotic regulation, cell wall integrity, and photoperiodic regulation and play critical roles in plant growth (Table 1). MYB and bHLH transcription factors together in an intricate order of various physiological processes in plants (Wang et al., 2022). Among these proteins, bHLH71 is a regulatory protein whose expression was highly changed in RhMYB24-silenced plants (Table 1, Figure 5A). Besides, RhMYB24 with RhbHLH71 showed strong transcription activity (Figure 4C). These results indicate that RhMYB24 directly binds with RhbHLH71 and promotes its transcription. These results are aligned with previous studies that MYB/bHLH work together and co-activate transcription activities of each other (Pireyre and Burow 2015).
Several bHLH proteins are influenced by auxin and play an important role in flower growth and development (Groszmann et al., 2008). We found the RhbHLH71 transcript was highly expressed in petals compared to other floral parts (Figure 5B). The transcript was high at the early stages of flower opening and went down when the flower fully opened (Figure 5C). Additionally, its transcripts are induced by Auxin and highly repressed by ethylene (Figure 5D), and flower opening was delayed in RhbHLH71-silenced plants compared to TRV control plants (Figure 6). In germinating seedlings, the cotyledon size is controlled by SPT a bHLH protein (Penfield et al., 2005). Several ARFs are important regulators of bHLH proteins. In Arabidopsis, 23 ARFs reported and divided into activating or repressing ARFs. Among the activating ARFs, ARF6-8 bond with bHLH proteins such as BEE2, HBI1, and PIF4, and are essential for cell elongation (Oh et al. 2014; Chandler 2016; Reed et al. 2018). The petal area reduced in RhbHLH71 silenced petals and the transcription level of different genes involved in cell expansion and cell wall integrity was downregulated. These results suggest that RhbHLH71 affects the petal’s size by regulating petal cell expansion. The mutation of SPT, a bHLH protein functioned in petal expansion that has big cell size (Penfield et al., 2005).
We observed RhMYB24 and RhbHLH71 mediate crosstalk of phytohormone-related genes that participated in petal cell expansion. The transcript level of auxin-related genes RhYUCCA6, RhIAA14, RhGH3, and Rh15A-like were lower in both RhMYB24 and RhbHLH71 silenced petals (Figure 8C, 8D). In the rose ‘Old Blush’, during petal expansion, it was found that genes related to auxin signaling such as SAUR, GH3, and AUX/IAA were upregulated and supported in flower opening (Han et al., 2019). In Arabidopsis, the ARF8 interacts with BPEp and controls the petal cell expansion by regulating the auxin-responsive genes Aux/IAAs1/3/9/17/19 (Varaud et al., 2011).
We observed that GAs biosynthesis gene RhGA20ox expression was downregulated, while GAs biosynthesis inhibited gene RhGA2ox expression upregulated in silenced plants (Figure 8A, 8B). We also found that an ethylene-regulated gene RhTIP1;1 promotes cell expansion its expression downregulated, while another ethylene-regulated protein that suppresses cell expansion RhGAI1, its expression was upregulated in silenced plants (Figure 8A, 8B). Earlier studies found that bHLH helps plants in osmotic regulation and flavonoid biosynthesis, hence keeping cells fully expanded and turgid (Qian et al., 2021). Ethylene can inhibit GA-induced root elongation in Arabidopsis (Achard et al., 2003). GAs activates their signaling pathway by inducing rapid degradation of DELLA proteins, which are GAs signaling repressors (Zhou et al., 2016), and RhHB1-RhGA20ox1 regulatory checkpoint controls the antagonistic effect between ethylene and GAs (Lv et al., 2014).
Overall, the discussion supports and signifies that RhMYB24 and RhbHLH71 form a complex and coordinate flower opening by mediating the crosstalk among phytohormones auxin, GAs, and ethylene-regulated genes, which participate in petal cell expansion. However, further studies are needed to find how auxin, GAs, and ethylene regulate RhMYB24 and RhbHLH71, and how these transcription factors regulate cell expansion genes.
Conclusion
In summary, our results advocate a possible model for the regulation of auxin and ethylene in flower openings. These hormones fine-tune the expression of RhMYB24 and RhbHLH71 to regulate cell expansion. The regulation of RhMYB24 and RhbHLH71 controlled genes related to auxin, GAs, and ethylene-regulated pathways (Supplementary Fig. S1). Our findings presented in this study provide a novel insight into RhMYB24 and RhbHLH71 coordinated flower opening through cell expansion genes by mediating the crosstalk between auxin and ethylene signaling. This study laid an important foundation for understanding the physiological process of flower opening and molecular breeding. It would help to maximize cut flowers’ ornamental and economic value.
Competing interests: The authors declare no competing interests.
Acknowledgments: The authors would like to acknowledge the support and guidance of the teachers and colleagues at the Department of Ornamental Horticulture, China Agricultural University, Beijing.
Reference
- Achard, P., Vriezen, W.H.,van der Straeten, D.,Harberd, N.P., 2003. Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell, 15: 2816–2825.
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C., 2007. Control of cell and petal morphogenesis by R2R3 MYB transcription factors, Development, 134: 1691-1701.
- Bey M, Stüber K, Fellenberg K, Schwarz-Sommer Z, Sommer H, Saedler H, Zachgo S., 2004. Characterization of antirrhinum petal development and identification of target genes of the class B MADS box gene DEFICIENS. Plant Cell 16: 3197-3215.
- Chandler JW., 2016. Auxin response factors. Plant Cell Environ 39: 1014–1028
- Chaumont, F.,Tyerman, S.D., 2014. Aquaporins: highly regulated channels controlling plant water relations. Plant Physiol, 164: 1600–1618.
- Chen C, Hussain N, Wang Y, Li M, Liu L, Qin M, Ma N, Gao J, Sun X., 2020. An Ethylene-inhibited NF-YC Transcription Factor RhNF-YC9 Regulates Petal Expansion in Rose. Horticultural Plant Journal 6, 419-427.
- Chen, C., Hussain, N., Ma, Y., Zuo, L., Jiang, Y., Sun, X., Gao, J., 2023. The ARF2-MYB6 module mediates auxin-regulated petal expansion in rose. Journal of Experimental Botany, 74(15), 4489–4502. https://doi.org/10.1093/jxb/erad173
- Chen, W.,Yin, X.,Wang, L.,Tian, J.,Yang, R.Y.,Liu, D.F.,Yu, Z.H.,Ma, N.,Gao, J.P., 2013. Involvement of rose aquaporin RhPIP1;1 in ethylene-regulated petal expansion through interaction with Rh- PIP2;1. Plant Mol Biol, 83: 219–233.
- Cheng C, Yu Q, Wang Y, Wang H, Dong Y, Ji Y, Zhou X, Li Y, Jiang CZ, Gan SS, Zhao L, Fei Z, Gao J, Ma N., 2021. Ethylene-regulated asymmetric growth of the petal base promotes flower opening in rose (Rosa hybrida). Plant Cell 33, 1229-1251.
- Cheng H, Qin L, Lee S, Fu X, Richards DE, Cao D, Luo D, Harberd NP, Peng J. 2004. Gibberellin regulates Arabidopsis floral development via suppression of DELLA protein function. Development 131, 1055-1064.
- Cosgrove, D.J., 2005. Growth of the plant cell wall. Nat Rev Mol Cell Biol, 6: 850–861.
- de Lucas, M.,Daviere, J.M.,Rodriguez-Falcon, M.,Pontin, M.,Igle- sias-Pedraz, J.M.,Lorrain, S.,Fankhauser, C.,Blazquez, M.A.,Titarenko, E.,Prat, S., 2008. A molecular framework for light and gibberellin control of cell elongation. Nature, 451: 480–484.
- Du M, Spalding EP, Gray WM., 2020. Rapid auxin-mediated cell expansion. Annual Review of Plant Biology 71, 379-402.
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L., 2010. MYB transcription factors in Arabidopsis. Trends Plant Sci. 15(10):573-81.
- Groszmann M, Paicu T, Smyth DR., 2008. Functional domains of SPATULA, a bHLH transcription factor involved in carpel and fruit development in Arabidopsis, The Plant Journal, 55: 40-52.
- Groszmann M, Bylstra Y, Lampugnani ER, Smyth DR., 2010. Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. Journal of Experimental Botany 61: 1495–1508.
- Han Y, Yong X, Yu J, Cheng T, Wang J, Yang W, Pan H, Zhang Q., 2019. Identification of candidate adaxial-abaxial-related genes regulating petal expansion during flower opening in Rosa chinensis “Old Blush”. Front Plant Sci 10, 1098.
- Hellens, R.P., Allan, A.C., Friel, E.N., Bolitho, K., Grafton, K., Templeton, M.D., Karunairetnam, S., Gleave, A.P., and Laing, W.A. (2005). Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13.
- Higginson, T., Li, S. F., & Parish, R. W., 2003. AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. The Plant Journal: for cell and molecular biology, 35(2), 177–192. https://doi.org/10.1046/j.1365-313x.2003.01791.x
- Ke M, Gao Z, Chen J, Qiu Y, Zhang L, Chen X., 2018. Auxin controls circadian flower opening and closure in the waterlily. BMC Plant Biology 18, 143.
- Li SB, Xie ZZ, Hu CG, Zhang JZ. 2016. A review of auxin response factors (ARFs) in plants. Front Plant Sci 7, 47.
- Luo J, Ma N, Pei HX, Chen JW, Li J, Gao JP., 2013. A DELLA gene, RhGAI1, is a direct target of EIN3 and mediates ethylene-regulated rose petal cell expansion via repressing the expression of RhCesA2. Journal of Experimental Botany 64, 5075-5084.
- Luo, J.,Ma, N.,Pei, H.X.,Chen, J.W.,Li, J.,Gao, J.P., 2013. A DELLA gene, RhGAI1, is a direct target of EIN3 and mediates ethylene-regulated rose petal cell expansion via repressing the expression of RhCesA2. J Exp Bot, 64: 5075–5084.
- Lv, P.T.,Zhang, C.Q.,Liu, J.T.,Liu, X.W.,Jiang, G.M.,Jiang, X.Q.,Khan, M.A.,Wang, L.S.,Hong, B.,Gao, J.P., 2014. RhHB1 mediates the antagonism of gibberellins to ABA and ethylene during rose Rosa hybrida petal senescence. Plant J, 78: 578–590.
- Ma N, Cai L, Lu W, Tan H, Gao J., 2005. Exogenous ethylene influences flower opening of cut roses (Rosa hybrida) by regulating the genes encoding ethylene biosynthesis enzymes. Sci China Ser C Life Sci 48, 434-444.
- Ma N, Xue JQ, Li YH, Liu XJ, Dai FW, Jia WS, Luo YB, Gao JP., 2008. Rh-PIP2;1, a rose aquaporin gene, is involved in ethylene-regulated petal expansion. Plant Physiology 148, 894-907.
- Meng, Y.,Li, N.,Tian, J.,Gao, J.,Zhang, C., 2013. Identification and validation of reference genes for gene expression studies in postharvest rose flower (Rosa hybrida). Sci Hortic, 158: 16–21.
- Nelson, B.K., Cai, X., and Nebenführ, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51: 1126–1136.
- Oh E., Zhu JY., Bai MY., Arenhart RA., Sun Y., Wang ZY., 2014. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3: e03031
- Pei H, Ma N, Tian J, Luo J, Chen J, Li J, Zheng Y, Chen X, Fei Z, Gao J., 2013. An NAC transcription factor controls ethylene-regulated cell expansion in flower petals. Plant Physiology 163, 775-791.
- Penfield S, Josse E-M, Kannangara R, Gilday AD, Halliday KJ, Graham IA., 2005. Cold and light control of seed germination through the bHLH transcription factor SPATULA, Current Biology, 15:1998-2006.
- Pireyre, M., & Burow, M., 2015. Regulation of MYB and bHLH transcription factors: a glance at the protein level. Molecular plant, 8(3), 378–388.
- Preston, J., Wheeler, J., Heazlewood, J., Li, S. F., & Parish, R. W., 2004. AtMYB32 is required for normal pollen development in Arabidopsis thaliana. The Plant Journal: for cell and molecular biology, 40(6), 979–995.
- Qian Y, Zhang T, Yu Y, Gou L, Yang J, Xu J, Pi E., 2021. Regulatory Mechanisms of bHLH Transcription Factors in Plant Adaptive Responses to Various Abiotic Stresses. Front Plant Sci. 12: 677611.
- Raymond, O.,Gouzy, J.,Just, J.,Badouin, H.,Verdenaud, M.,Lemainque, A.,Vergne, P.,Moja, S.,Choisne, N.,Pont, C.,Carrere, S.,Caissard, J.C.,Couloux, A.,Cottret, L.,Aury, J.M.,Szecsi, J.,La- trasse, D.,Madoui, M.A.,Francois, L.,Fu, X.P.,Yang, S.H.,Dubois, A.,Piola, F.,Larrieu, A.,Perez, M.,Labadie, K.,Perrier, L.,Govetto, B.,Labrousse, Y.,Villand, P.,Bardoux, C.,Boltz, V.,Lopez-Roques, C.,Heitzler, P.,Vernoux, T.,Vandenbussche, M.,Quesneville, H.,Boualem, A.,Bendahmane, A.,Liu, C.,Le Bris, M.,Salse, J.,Baudino, S.,Benhamed, M.,Wincker, P.,Bendahmane, M., 2018. The Rosa genome provides new insights into the domestication of modern roses. Nat Genet, 50: 772–777.
- Reed JW, Wu MF, Reeves PH, Hodgens C, Yadav V, Hayes S, Pierik R (2018) Three auxin response factors promote hypocotyl elongation. Plant Physiol 178: 864–875
- Rose, J.K.C.,Braam, J.,Fry, S.C.,Nishitani, K., 2002. The xth family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomencla- ture. Plant Cell Physiol, 43: 1421–1435.
- Shin, B., Choi, G., Yi, H., Yang, S., Cho, I., Kim, J., Lee, S., Paek, N. C., Kim, J. H., Song, P. S., & Choi, G., 2002. AtMYB21, a gene encoding a flower-specific transcription factor, is regulated by COP1. The Plant journal: for cell and molecular biology, 30(1), 23–32.
- Strader LC, Zhao Y. 2016. Auxin perception and downstream events. Current Opinion in Plant Biology 33, 8-14.
- Sun X, Qin M, Yu Q, Huang Z, Xiao Y, Li Y, Ma N, Gao J., 2021. Molecular understanding of postharvest flower opening and senescence. Molecular Horticulture 1, 7.
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M., 2006. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. EMBO Journal 25, 3912-3920.
- Tian, J.,Pei, H.X.,Zhang, S.,Chen, J.W.,Chen, W.,Yang, R.Y.,Meng, Y.L.,You, J.,Gao, J.P.,Ma, N., 2014. TRVGFP: a modified Tobacco rattle virus vector for efficient and visualizable analysis of gene function. J Exp Bot, 65: 311–322.
- van Doorn WG, Dole I, Celikel FG, Harkema H., 2013. Opening of Iris flowers is regulated by endogenous auxins. Journal of Plant Physiology, 170, 161-164.
- Varaud E, Brioudes F, Szécsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M., 2011. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23, 973-983.
- Verdoucq, L.,Rodrigues, O.,Martiniere, A.,Luu, D.T.,Maurel, C., 2014. Plant aquaporins on the move: reversible phosphorylation, lateral motion, and cycling. Curr Opin Plant Biol, 22: 101–107.
- Wang B, Luo Q, Li Y, Du K, Wu Z, Li T, Shen WH, Huang CH, Gan J, Dong A., 2022. Structural insights into partner selection for MYB and bHLH transcription factor complexes. Nat Plants. 8(9):1108-1117.
- Wang J, Wang H, Ding L, Song A, Shen F, Jiang J, Chen S, Chen F., 2017. Transcriptomic and hormone analyses reveal mechanisms underlying petal elongation in Chrysanthemum morifolium ‘Jinba’. Plant Molecular Biology 93, 593-606.
- Wu L, Ma N, Jia YC, Zhang Y, Feng M, Jiang CZ, Ma C, Gao JP., 2017. An ethylene-induced regulatory module delays flower senescence by regulating cytokinin content. Plant Physiology 173, 853-862.
- Xue, J.Q., Li, Y.H., Tan, H., Yang, F., Ma, N., and Gao, J., 2008. Expression of ethylene biosynthetic and receptor genes in rose floral tissues during ethylene-enhanced flower opening. J Exp Bot, 59: 2161-2169.
- Yamada K, Norikoshi R, Suzuki K, Nishijima T, Imanishi H, Ichimura K., 2009. Cell Division and Expansion Growth during Rose Petal Development. Journal of the Japanese Society for Horticultural Science, 78, 356-362.
- Yang C, Xu Z, Song J, Conner K, Vizcay Barrena G, Wilson ZA. 2007. Arabidopsis MYB26/MALE STERILE35 regulates secondary thickening in the endothecium and is essential for anther dehiscence. Plant Cell 19, 534-548.
- Zhang S, Feng M, Chen W, Zhou X, Lu J, Wang Y, Li Y, Jiang CZ, Gan SS, Ma N, Gao J., 2019. In rose, transcription factor PTM balances growth and drought survival via PIP2;1 aquaporin. Nat Plants 5, 290-299.
- Zhang, H., Liang, W., Yang, X., Luo, X., Jiang, N., Ma, H., & Zhang, D., 2010. Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. The Plant Cell, 22(3), 672–689.
- Zhou, X., Zhang, Z.L., Park, J., Tyler, L., Yusuke, J., Qiu, K., Nam, E.A., Lumba, S., Desveaux, D., McCourt, P., Kamiya, Y., Sun, T.P., 2016. The ERF11 transcription factor promotes internode elongation by activating gibberellin biosynthesis and signaling. Plant Physiol, 171: 2760–2770.
- Zhu, X., Liang, W., Cui, X., Chen, M., Yin, C., Luo, Z., Zhu, J., Lucas, W. J., Wang, Z., and Zhang, D., 2015. Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. The Plant journal: for cell and molecular biology, 82(4), 570–581. https://doi.org/10.1111/tpj.12820
Supplementary Data:
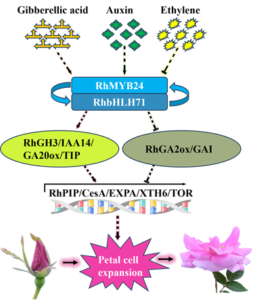
Supplementary Fig. S1. Schematic diagram of RhMYB24 and RhbHLH71 coordinated flower opening.
RhMYB24 and RhbHLH71 were induced by auxin and inhibited by ethylene. They interact together in a complex and control petal size through cell expansion-related genes. RhMYB24 and RhbHLH71 mediate the crosstalk between auxin and ethylene signaling by regulating genes related to auxin, GAs, and ethylene that control flower openings.
| Supplementary Table 1. Primers List | ||
| Gene | Primers Sequence (5′ -3′) | |
| RhUBI2 | F | GCCCTGGTGCGTTCCCAACTG |
| R | CCTGCGTGTCTGTCCGCATTG | |
| RhMYB24 | F | CAAGTGGGGAAACAGGTGGT |
| R | GACGATGAGTTGGGGGTGTT | |
| TRV-MYB24 | F | GAATTCCAAGTGGGGAAAC |
| R | GGATCCGACGATGAGTTGG | |
| RhCesA2 | F | GACTTGCTACGAGTATGAGCG |
| R | TATGGAAATCGGAGGAGACCG | |
| RhPIP1;1 | F | CAAGAGGGCACCAAACATGTG |
| R | CTCAAACCCCTTGACCACAG | |
| RhPIP2;1 | F | CTCTCATAGCCGAGTTCATCG |
| R | GATCAGAGAGACCTTACGAGC | |
| RhEXPA4 | F | TAGAGGTAGACCGAGTTGGAT |
| R | TGTCCACAAGTGGCAGCAAGA | |
| RhEXPA8 | F | ATGGCAGCTTCAGCATTGTC |
| R | CCTCATTTCATAGCAAGAGCC | |
| RhXTH6 | F | CCAAGTCACATGGTCTGATTC |
| R | GTACTGTATCTGTGTCCGAG | |
| RhTOR3 | F | GACTAATCTCGAGTCGCGCA |
| R | CCGAAACCTTCGGTTCTCCA | |
| RhTOR4 | F | CACCTGACACCTCCCATCAC |
| R | GTCACGGTCGGAGAGTTTGT | |
| RhbHLH71 | F | GAAGCAGCTGCCTTGTGTTA |
| R | AGATTAACGCGATTCTGGACTCA | |
| TRV-bHLH71 | F | GAATTCGAAGCAGCTGCCTT |
| R | GGATCCAGATTAACGCGATTCTG | |
| RhYUCCA6 | F | AGATGGGTTGCCAAAAACGC |
| R | CCATCACCAAAACCCTCCGA | |
| RhIAA14 | F | AAACGTCCCACGCAGTGATA |
| R | GCCTGTGAATGTCCGTATGC | |
| RhGH3 | F | CGGGCCACTACGTCATCTAC |
| R | GGGGTCCAATGCGGTAAAGA | |
| Rh15A-like | F | TCCAGCTTGAGTGTGTGACT |
| R | ACATGGGCTGCATACAGAAT | |
| RhGA20ox | F | CTGTCCTCCTTGCTAACTCATCCTC |
| R | CTCGTATTGAAGAACTGGAGCATCG | |
| RhGA2ox | F | TGGGCCAAGGAATGTGTTCA |
| R | GGGTCTGTGTGCTCTCCAAA | |
| RhTIP1;1 | F | GAGCTGGAACTGGGAGAACC |
| R | TCATAAACAAGCCCGGCGAT | |
| RhGAI1 | F | ACCACCCTAATCCAAACCCTTC |
| R | AGCCCGTCTTCCTGAACTGTC | |
| TRV1 | F | TTACAGGTTATTTGGGCTAG |
| R | CCGGGTTCAATTCCTTATC | |
| TRV2 | F | TGGGAGATGATACGCTGTT |
| R | CCTAAAACTTCAGACACG | |