Publication Information
ISSN: 2641-6816
Frequency: Continuous
Format: PDF and HTML
Versions: Online (Open Access)
Year first Published: 2018
Language: English
| Journal Menu |
| Editorial Board |
| Reviewer Board |
| Articles |
| Open Access |
| Special Issue Proposals |
| Guidelines for Authors |
| Guidelines for Editors |
| Guidelines for Reviewers |
| Membership |
| Fee and Guidelines |
 |
The Murine Major Facilitator Superfamily Domain Containing 14A (Mfsd14a) Gene Does Not Encode a Glucose Transporter
Zhouyao H1, Fehsenfeld S2, 3, Weihrauch D3 & Eck K P1*
1University of Manitoba, Department of Food and Human Nutritional Sciences, 190 Dysart Road, Winnipeg, MB R3T 2N2, Canad
2Université du Quebec à Rimouski, Département de biologie, chimie et géographie, 300 Allée des Ursulines, Rimouski, QC G5L 3A1, Canada
|3University of Manitoba, Department of Biological Sciences, 190 Dysart Road, Winnipeg, MB R3T 2N2, Canada
Received Date: February 04, 2022; Accepted Date: February 16, 2022; Published Date: February 22, 2022;
*Corresponding Author: Peter Karl Eck, University of Manitoba, Department of Food and Human Nutritional Sciences, 66 Chancellors Cir, Winnipeg, MB R3T 2N2, Canada. Email: Peter.Eck@umanitoba.ca
Citation: Zhouyao H, Fehsenfeld S, Weihrauch D & Eck K P (2022) The Murine Major Facilitator Superfamily Domain Containing 14A (Mfsd14a) Gene Does Not Encode a Glucose Transporter. Adv in Nutri and Food Sci: ANAFS-224.
DOI: 10.37722/ANAFS.2022101
Abstract
Despite that fact that the mfsd14a knock-out mouse shows a phenotype resembling human globozoospermia, the literature record on the mouse mfsd14a gene and its encoded products is extremely limited. This report therefore aims to establish the baseline on the mouse genomic locus.
Utilizing visual supervised bioinformatics of the mfsd14a genomics locus and its encoded products are annotated. Our analysis revealed a single protein isoform with all the hallmarks of a functional membrane bound solute carrier. The gene’s sole transcript is almost ubiquitously expressed with highest expression in reproductive tissues followed by nervous tissues. Despite previous suggestions, the murine mfsd14a protein does not mediate transmembrane glucose transport when expressed in Xenopus laevis oocytes.
Introduction
The public literature record on the mouse mfsd14a gene, its encoded protein products and function(s) is virtually non existent. Only six credible publications can be identified in PubMed or in Scopus referring to murine mfsd14a, none of them identifying the encoded product and its function (Doran et al., 2016; Lekholm et al., 2017; Luco et al., 2012; Matsuo et al., 1997; S. Sreedharan et al., 2011; Wang et al., 2017).
Two publications provide very limited information about the gene’s transcript in murine species. Specifically, Matsu et al. (1997) cloned and sequenced the gene transcript (cDNA) from neonatal mouse hippocampus and named it “hiat1 (hippocampus abundant transcript 1)”. The authors predicted that the encoded product is a transmembrane protein. Based on sequence similarities of the predicted protein and the presence of “signature motifs” (supplemental information Figure 1), the authors further extrapolated that the mouse mfsd14a might encode for a novel sugar transporter (Matsuo et al., 1997), however, no functional data were provided.
Matsu et al. (1997) showed gene expression of the murine mfsd14a in almost all mouse tissues they investigated, which was mirrored by Sreedharan et al. (2011) for tissues examined in rat (Smitha Sreedharan et al., 2011). However, both studies did not provide details on standardized expression levels and only examined a limited spectrum of tissues.
Altered spermatogenesis and male infertility is observed in mfsd14a-/- mice (Doran et al., 2016), where homozygous mutant mice were viable and healthy, but males were sterile due to a 100-fold reduction in the number of spermatozoa in the vas deferens. The few spermatozoa that were formed showed rounded head defects like those found in humans with globozoospermia. The expression of mfsd14a in Sertoli cells (“nurse cells”) led Doran and coworkers to suggest that mfsd14a may transport a nutrient or metabolite from the bloodstream into the cells that is required for spermatogenesis.
Lekholm and coworkers described mfsd14a gene expression in the mouse central nervous system, and the location of the protein in the Golgi system and endoplasmatic reticulum (Lekholm et al., 2017). Based on the differential regulation data, the authors also predicted an organic substrate with a possible involvement in energy homeostasis, such as glucose. This was deduced because an upregulation was reported after 3 hours of amino acid starvation in mouse embryonic primary cultures. By contrast, in mice subjected to 24 hours of starvation a downregulation of mfsd14a was shown in the hypothalamus and the brainstem. Moreover, a high fat diet caused upregulation in the striatum (Lekholm et al., 2017).
The emerging disease associations precipitate the need for a detailed annotation of the mouse mfsd14a locus and a characterization of its predicted function as a putative glucose transporter. Based on visually supervised analysis of available data we annotate mouse mfsd14a and its encoded products. Its functions as a solute carrier for glucose is interrogated in the heterologous Xenopus laevis oocytes expression system.
Material and Methods
Bioinformatics genomics analysis
The National Center for Biotechnology Information (NCBI) database was used to obtain the genomic sequence (Chromosome 3 – NC_000069.7), reference, nonreference full length RNA, and the expressed sequence tags (ESTs) sequences for mouse mfsd14a. The DNA sequence analysis software Sequencher 4.8 (GeneCodes Corporation; Ann Arbor, Michigan, USA) was used to create alignments between the reference RNA sequence and the genomic sequence to determine the structures of exons and introns. The alignment parameter used were large gaps, 20 minimum overlap, and minimum match of 80%. These alignments were visually curated to determine exons and introns and to create the scaled illustrations.
Protein analysis
Protein Homology/analogY Recognition Engine V 2.0 (Phyre2, (Kelley et al., 2015)) and the AlphaFold Model using 3D ligand site (Jumper et al., 2021; Wass et al., 2010) were used to analyse the function and structure of the mfsd14a protein. During the analysis, parameters from the standard/normal modes were applied.
Expression patterns analysis
Experimental RNA-Seq data were interrogated at the following resources: Gene Expression Database (GXD), Mouse Genome Informatics Web Site. World Wide Web (URL: http://www.informatics.jax.org/expression.shtml) (January 23, 2022); European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) Baseline Expression Atlas (URL: https://www.ebi.ac.uk/gxa/home) (Moreno et al., 2022) and The Encyclopedia of DNA Elements (ENCODE) (URL: https://www.encodeproject.org, (Consortium, 2012)).
Plasmid preparation
Mouse fd14a. Initially, the complete open reading frame (ORF; 1701 bp) for the mouse mfsd14a (GenBank accession no. NM_008246.2) was amplified from mouse brain cDNA with Phusion high fidelity DNA polymerase (New England Biolabs, Ipswich, Massachusetts, USA) and subcloned into the p426 expression vector for amplification in yeast applying sticky-end ligation with T4 ligase (New England Biolabs, Ipswich, Massachusetts, USA) using the restriction enzymes SpeI and SmaI (please see Table 1 in supplemental information for sequence details) Subsequently, the ORF was sent for sequencing at the DNA Sequencing Facility of the Robarts Research Institute (London, Ontario, Canada) using T7/SP6 primers. For functional expression in Xenopus laevis oocytes, the verified mfsd14a ORF was then subcloned from the p426 vector into the pGEM®-HE plasmid, a modified pGEM®-3Z vector for the cloning site to be flanked by Xenopus beta globin 5’- and 3’- UTR sequence, using T4 ligase as described above. Both primers for this subcloning contained a restriction site for SmaI (forward primer including T7/SmaI-restriction site/ATG and reverse primer containing SmaI-restriction site). The insertion and the correct orientation of the ORF was verified by PCR (T7 plus mfsd14a internal reverse primer) and subsequent sequencing with T7/SP6 primers (supplied by the Centre for Applied Genoics (TCAG) sequencing facility, Toronto, Canada).
Human GLUT3. Plasmid containing the full open reading frames (ORF) human GLUT3 (positive control for glucosetransport study) was purchased from Harvard PlasmID (clone ID: HsCD00021270) and transformed into DH5α competent cells (New England Biolabs, Ipswich, Massachusetts, USA) following the manufactures guidance for propagation, followed by column purification (QIAprep Spin Miniprep Kit, Qiagen, Hilden, Germany). The ORFs were then amplified with Q5 High Fidelity DNA Polymerase (New England Biolabs) and was tagged with T7 RNA polymerase promoter sequence. The GLUT3 amplicon containing GLUT3 ORF and T7 promoter sequence was used directly for cRNA synthesis as described previously. cRNA products were column purified (RNeasy MinElute Cleanup Kit, Qiagen) prior to the spectrophotometric quantification (Nanodrop, ND-1000, Thermofisher, Waltham, MA, USA) and the integrity assessments (MOPS agarose gel containing formaldehyde).
Functional expression of mfsd14a in Xenopus laevis oocytes
Chemicals and reagents. [3H] 2-deoxyglucose (25.5 Ci/mmol in water) was purchased from PerkinElmer (Waltham, MA, USA). All chemicals were purchased from Sigma-Aldrich (St. Louis, MI, USA) unless otherwise noted. The standard oocyte ringer (OR2) contained (in mmol L-1) 82.5 NaCl, 2.5 KCl, 1 CaCl2, 1 MgCl2, 1 Na2HPO4, 5 HEPES, pH 7.4 and sterilized by vacuum bottle-top filters (EMD MilliporeTM SteritopTM sterile vacuum bottle-top filters, ThermoFisher, Waltham, Massachusetts, USA). After sterilization, OR2 was then supplemented with sodium pyruvate (2.5 mmol L-1), penicillin-streptomycin (1 mg mL-1) (Gibco, Long Island, NY, USA), and gentamicin (50 μg mL-1) for long term storage of sorted oocytes.
Plasmid preparation. The pGEM®-HE plasmid containing the ORF for mfsd14a as described above, was transferred into DH5-alpha cells for amplification, column purified (Plasmid miniprep kit, Qiagen, Hilden, Germany) and linearized with SphI (New England Biolabs (NEB), Ipswich, Massachusetts, United States). The HiScribe™ T7 ARCA mRNA Kit (with tailing) (NEB) was used for the in-vitro transcription of the capped mRNA (cRNA) following the manufacturer’s suggestion, column purified (RNeasy MinElute Cleanup Kit, Qiagen, Hilden, Germany) and eluted in nuclease free water.
Oocyte preparation. Stage VI-V oocytes were collected from mature female Xenopus laevis (VWR International, Randor, PA, USA) as previously described (Soreq, 1992). Briefly, the frogs were anesthetized with MS-222 (2 g L-1) followed with decapitation prior to the collection of the ovary. The ovary was placed in Ca2+-free OR2 solution containing collagenase type VI (1 mg mL-1) (Gibco, Waltham, Massachusetts, USA) while gently agitated for 90 minutes at room temperature. The activity of collagenase was terminated by rinsing the oocytes three times with standard OR2. Oocytes were then sorted manually, rinsed three additional times with standard OR2 and allowed to recover in sterile OR2 overnight at 16 °C. All procedures used were approved by the University of Manitoba Animal Research Ethics Board and are in accordance with the Guidelines of the Canadian Council on Animal Care.
Microinjections of oocytes. After recovering overnight, 18.4 ng (36.8 nL with 0.5 ng nL-1) of mfsd14a cRNA, human glucose transporter 3 (GLUT3) cRNA (as positive control), or nuclease-free water (as negative control, sham) was delivered into the oocytes using the Nanoject II auto-nanoliter injector (Drummond Scientific, Broomall, Pennsylvania, USA). The glucose uptake experiments were done at room temperature two days after the injections, where oocytes were randomly divided into groups containing 20 oocytes each either injected with water (sham) or cRNA (mfsd14a or GLUT3). Oocytes were incubated in 200 μL standard OR2 with 125 pmol [3H] 2-deoxyglucose for 30 minutes. Excess ice-cold standard OR2 was used terminate the transport, add was used to rinse oocytes four times to remove the external radioactivity. Oocytes were then lysed individually in 200 μL of 10% sodium dodecyl sulfate (SDS) after which 5 ml Ultima Gold scintillation cocktail (PerkinElmer) was added. Internal radioactivity was quantified by liquid scintillation spectrometry (Tri-Carb 2900 TR; PerkinElmer) as counts per minute (CPM)/oocyte.
The washing efficiency was ensured by assessing 50 μL of the radioactivity of the buffer collected from the fourth washing post radioactive exposure compared to fresh, radioactivity-free standard OR2 at the same volume. If excessive amount of radioactivity was found, the oocytes were discarded.
Statistical analysis
Data was tested for normal distribution and homogeneity of variances with the Bartlett’s test and one way ANOVA test with Tukey’s post hoc analysis were used to compare multiple means in the glucose uptake studies. All results with P < 0.05 were considered significant. Error bars indicate standard error of the mean (SEM) and different letters indicates p values less or equal to 0.05. Graphs were generated using the software GraphPad/Prism 9 for Mac OS, GraphPad Software, San Diego, California USA, www.graphpad.com).
Results
The mouse mfsd14a gene locus
The mouse mfsd14a gene is listed in NCBI as gene ID 15247 and resides on the mouse chromosome 3qG1, reverse strand. It is immediately flanked by the Spindle assembly abnormal protein 6 homolog (sass6) and the mir3473h and solute carrier family 35 member a3 (slc35a3) genes (Figure 1).
The visually supervised alignment of transcripts shows that the mouse mfsd14a gene spans 31,496 bases from its transcription initiation to the termination site. The transcript starts 11 nucleotides upstream of the site currently identified for the NCBI Reference Transcript NM_008246 (supplemental information Figure 2). In the terminal 3’ exon, two potential transcriptional termination sites are visualized (supplemental information Figure 3).
 Figure 1: The mouse mfsd14a genomic locus. The transcripts’ location on Chromosome 3 is indicated by the black arrow in the top panel. The exon-intron structure of the sole transcript is depicted on the bottom panel.
Figure 1: The mouse mfsd14a genomic locus. The transcripts’ location on Chromosome 3 is indicated by the black arrow in the top panel. The exon-intron structure of the sole transcript is depicted on the bottom panel.
The mouse mfsd14a protein
The mouse mfsd14a protein consists of 490 amino acids, as represented by NCBI reference protein NP_032272.2 (supplemental information Sequence 1). The protein is predicted to be a solute membrane carrier with twelve transmembrane domains (Figure 2A & B and Supplemental information Figure 4).
The mfsd14a protein clusters to the conserved protein domain family MFS, subfamily MFSD14, which contains only one additional member, the orthologous mfsd14b (Supplemental information Figure 5). A unique putative chemical substrate binding pocket is predicted. As putative organic substrates, glucose, xylose, maltose, alanine, chloramphenicol, and 3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID are predicted to bind in a pore-like intramolecular binding pocket (Figure 2C). This corroborates the speculations that glucose might be a main substrate.

Figure 2: The predicted secondary (panel A) and tertiary (panel B) structures for the mouse mfsd14a protein (Image colored by rainbow N→C terminus; Model dimensions (Å): X:58.386 Y:50.944 Z:54.127). Models created using the Protein
Homology/Analogy Recognition Engine V 2.0 (Phyre2) web-server (Kelley et al., 2015). Panel C depicts the putative ligand binding pocked aligned in a “pore-like” structure created as an AlphaFold Model using the 3D ligand site (Wass et al., 2010). The ligands are coloured in blue and the binding residues are coloured in red; the remaining protein structure is coloured in grey.
Tissues expression of the mouse mfsd14a transcript
mfsd14a tissue expression is almost ubiquitous, with highest expression in the testis, oocyte, corpus callosum, spermatid, thymus, mesenteric adipose tissue, brainstem, ovary (Figure 3 and Supplemental information Figures 6 and 7). Depending on the project analysed, almost all interrogated tissues showed some degree of expression (Figure 3 and Supplemental information Figures 6 and 7). Lowest expression was observed in muscle tissues.
High expression in the mouse central nervous system was reported. A more detailed interrogation shows a very distinct mfsd14a’s spatial distribution, located to single cell structures in the Main Olfactory Bulb inner plexiform layer, the pyramidal cell layer of the Hippocampal Formation, and the cerebellum ganglionic layer or Purkinje cell layer (Supplemental information Figure 8).

Figure 3: Mouse mfsd14a expression data (displayed as Reads Per Kilobase Million, RPKM) from the ENCODE project. Data were obtained via NCBI gene page, Gene ID: 15247.
Mouse mfsd14a does not mediate glucose uptake into Xenopus laevis oocytes.
mfsd14a expressing Xenopus laevis oocytes did not mediate the uptake of radiolabeled 2-deoxyglucose (Figure 4). Prior predictions that murine mfsd14a could mediate sugar transport (Matsuo et al., 1997), and speculation based on the expression sites that it could be a glucose transporter on the plasmalemma membrane are not supported by these experiments in Xenopus laevis oocytes.

Figure 4: The uptake of radiolabeled 2-deoxyglucose into Xenopus laevis oocytes expressing a known glucose carrier, human GLUT3 (clone ID: HsCD00021270) and mouse mfsd14a. N=15-20 oocytes per group.
Discussion
The orphan mfsd14a gene is currently underreported, specifically with respect to the mfsd14a-/- mouse phenotype resembling human globozoospermia. This report establishes the baseline annotation for the mouse gene and encoded products. Significantly, we identified one sole transcript to encode for one protein isoform, which is predicted to be a novel glucose membrane transporter both in the literature and with current bioinformatics tools.
However the mouse mfsd14a did not mediate uptake of glucose, the canonical substrate of many members of the slc2a (glut) and slc5a (sglt) family of transporters (Deng & Yan, 2016; Holman, 2020). The lack of glucose transport does not necessarily rule out transport of alternative saccharoses, but should emphasise the need to look beyond sugars as substrates. Supporting the potential for alternative substrates is the fact that a decrease in mRNA abundance of the mfsd14a paralogue in the gills of the European green crabs, Carcinus maenas, had been observed in response to elevated environmental pCO2 (Fehsenfeld et al., 2011). This observation might provide an avenue to explore further substrate candidates involved in acid-base homeostasis and regulation.
About 95% of human multi-exonic genes are alternatively spliced and there are in access of 100,000 alternative splicing events in major human tissues (Pan et al., 2008). Changes in splicing are known to affect the function and regulation of genes (Mazin et al., 2018), but splicing aberrations negatively correlate with fitness costs (Saudemont et al., 2017). Therefore, with no identified splice variants, the mouse mfsd14a gene can be considered to be under significant selective pressure resulting in high conservation in regard to splicing, but also in regard to nucleotide and amino acid conservation (Luo et al., 2015). The high conservation further corroborates that mfsd14a might have an essential nutrient/metabolite as a substrate, or to be a key protein in detoxifications or acid-base balance.
Mfsd14a’s widespread tissue expression also indicates a role in pathways critical for cell viability. However, the only deleterious phenotype reported for the mfsd14a-/- mouse is male infertility. This might be owed to the fact that the mfsd14a-/- mouse had not been fully phenotyped and had not been subjected to metabolic challenges reported to result in differential regulation in the brain (Lekholm et al. (2017). Further exploration of the mfsd14a-/- mouse model might reveal phenotypes related to brain functioning, since expression levels in brain tissues is comparable to testis. However, considering mfsd14a’s distinct spatial expression (Supplemental information Figure 8), existing reports on differential regulation in mouse brain under nutrient deprivation and in high fat diets, could be viewed with caution, since tissue isolation might have contributed to the observed differential regulation. Those experimental problems could be resolved utilizing single cell RNA sequencing technology in future experiments.
Conclusion
The mouse mfsd14a- gene is annotated to express a single transcript encoding one protein and is almost ubiquitously expressed. It does not mediate glucose uptake into Xenopus laevis oocytes. The genomic and functional baseline to explore disease phenotypes is established.
References
- Consortium E P (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489:57-74.
- Deng D & Yan N (2016). GLUT, SGLT, and SWEET: Structural and mechanistic investigations of the glucose transporters. Protein Sci, 25:546-558.
- Doran J, Walters C, Kyle V, Wooding P, Hammett-Burke R, et al. (2016). Mfsd14a (Hiat1) gene disruption causes globozoospermia and infertility in male mice. Reproduction, 152:91-99.
- Fehsenfeld S, Kiko R, Appelhans Y, Towle D W, Zimmer M, et al. (2011). Effects of elevated seawater pCO (2) on gene expression patterns in the gills of the green crab, Carcinus maenas. BMC Genomics, 12, 488.
- Holman G D (2020). Structure, function and regulation of mammalian glucose transporters of the SLC2 family. Pflugers Arch, 472:1155-1175.
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596:583-589.
- Kall L, Krogh A, Sonnhammer E L (2004). A combined transmembrane topology and signal peptide prediction method. J Mol Biol, 338:1027-1036.
- Kelley L A, Mezulis S, Yates C M, Wass M N, Sternberg M J (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc, 10:845-858.
- Krogh A, Larsson B, von Heijne G, Sonnhammer E L (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol, 305:567-580.
- Lekholm E, Perland E, Eriksson M M, Hellsten S V, Lindberg F A, et al. (2017). Putative membrane-bound transporters MFSD14A and MFSD14B are neuronal and affected by nutrient availability. Frontiers in Molecular Neuroscience, 10.
- Luco S, Delmas O, Vidalain P O, Tangy F, Weil R, et al. (2012). RelAp43, a Member of the NF-κB Family Involved in Innate Immune Response against Lyssavirus Infection. PLoS Pathogens, 8(12), Article e1003060.
- Luo H, Gao F & Lin Y (2015). Evolutionary conservation analysis between the essential and nonessential genes in bacterial genomes. Sci Rep, 5, 13210.
- Matsuo N, Kawamoto S, Matsubara K, Okubo K (1997). Cloning of a cDNA encoding a novel sugar transporter expressed in the neonatal mouse hippocampus. Biochemical and Biophysical Research Communications, 238:126-129.
- Mazin P V, Jiang X, Fu N, Han D, Guo M, et al. (2018). Conservation, evolution, and regulation of splicing during prefrontal cortex development in humans, chimpanzees, and macaques. RNA, 24:585-596.
- Moreno P, Fexova S, George N, Manning J R, Miao Z, et al. (2022). Expression Atlas update: gene and protein expression in multiple species. Nucleic Acids Res, 50:D129-D140.
- Pan Q, Shai O, Lee L J, Frey B J, Blencowe B J (2008). Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet, 40:1413-1415.
- Peters C, Tsirigos K D, Shu N, Elofsson A (2016). Improved topology prediction using the terminal hydrophobic helices rule. Bioinformatics, 32:1158-1162.
- Reynolds S M, Kall L, Riffle M E, Bilmes J A, Noble W S (2008). Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput Biol, 4:e1000213.
- Saudemont B, Popa A, Parmley J L, Rocher V, Blugeon C, et al. (2017). The fitness cost of mis-splicing is the main determinant of alternative splicing patterns. Genome Biol, 18:208.
- Soreq H, Seidman S (1992). Xenopus oocyte microinjection: from gene to protein. Methods in Enzymology, 207, 225-265.
- Sreedharan S, Stephansson O, Schioth H B, Fredriksson R (2011). Long evolutionary conservation and considerable tissue specificity of several atypical solute carrier transporters. Gene, 478:11-18.
- Viklund H, Elofsson A (2004). Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci, 13:1908-1917.
- Viklund H, Elofsson A (2008). OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics, 24:1662-1668.
- Wang K, Sun Y, Tao W, Fei X, Chang C (2017). Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett. 394:1-12.
- Wass M N, Kelley L A, Sternberg M J (2010). 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res, 38:W469-473.
Supplement Information:
Supplemental information Table 1: Sequences of primers used in the cloning process of mouse mfsd14a.
Lower case letters indicate restriction enzyme recognition sites.
Name
Sequence 5'à 3'
mmfsd14a SpeI forward
ATAactagtATGACCCAGGGGAAGAAA
mmfsd14a SmaI reverse
ATAcccgggAGTGGCAAGAGAGTGGTGCT
mmfsd14a SmaI T7 forward
TAATACGACTCACTATAGGGcccgggATGAAGAACAGAGTTGCAGG
mmfsd14a SmaI reverse 2
AATcccgggAGTGGCAAGAGAGTGGTGCT.
mmfsd14a T7 verification forward
AGGGTGTTGTTCAAGGGAT
mmfsd14a internal reverse
AGTGGCAAGAGAGTGGTGCT
GLUT3 ORF forward
ATGGGGACACAGAAGGTCACCCCAG
GLUT3 ORF reverse
TTAGACATTGGTGGTGGTCTCCTTA
GLUT3 T7 forward
AAAATAATACGACTCACTATAGACCATGGGGACACAGAAGGTCAC

Supplemental information Figure 1: signature motifs in the mouse mfsd14a (hiat1) protein aligning with known monosaccharide transporters, as depicted in Matsuo et al. (1997, Figure 3). The D-R/K-X-G-R-R/K motif shown in panel A and the P-E-S-P-R motif shown in panel B of mouse mfsd14a (formally known as HiAT1, or hippocampus abundant gene transcript 1), tetracycline efflux major facilitator superfamily transporter C (Tet C), the human tetracycline transporter-like protein (Tet-like), mouse glucose transporter type 2 (GLUT2) and mouse glucose transporter type 4. Homologous regions are shaded, and the putative transmembrane domains (TM) are boxed. Numbers on the left indicates amino acid numbers.
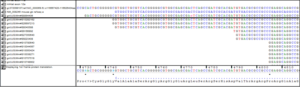
Supplemental information Figure 2: Alignment of the 5’ transcripts (EST and refence sequence evidence form NCBI) in the mouse mfsd14a locus. EST Mm#S8894406 (marked with a black line) shows eleven additional 5’ nucleotides currently not included in the reference sequence NM_008246.
 Supplemental information Figure 3: An overview of the alignments of the 3’ transcripts (EST and refence sequence evidence form NCBI) in the mouse mfsd14a locus. Green arrows represent EST sequences sequenced from the 5’ to 3’ direction and the red arrows represent EST sequences sequenced from the 3’ to 5’ direction. After all the available EST sequences were downloaded, they were aligned and visually curated to ensure the quality and accuracy of the alignment. Two transcription termination sites (also called poly a-tails, marked as arrow AA1 and AA2) are visualised. The most 5’ site does not impact on the protein coding region.
Supplemental information Figure 3: An overview of the alignments of the 3’ transcripts (EST and refence sequence evidence form NCBI) in the mouse mfsd14a locus. Green arrows represent EST sequences sequenced from the 5’ to 3’ direction and the red arrows represent EST sequences sequenced from the 3’ to 5’ direction. After all the available EST sequences were downloaded, they were aligned and visually curated to ensure the quality and accuracy of the alignment. Two transcription termination sites (also called poly a-tails, marked as arrow AA1 and AA2) are visualised. The most 5’ site does not impact on the protein coding region.
Supplemental information Sequence 1: Amino acid sequence of the full mouse Mfsd14a protein (formally known as hippocampus abundant transcript 1, Hiat1)
>NP_032272.2, Mfsd14a, formerly know as hippocampus abundant transcript 1 protein [Mus musculus]
MTQGKKKKRAANRSIMLAKKIIIKDGGTPQGIGSPSVYHAVIVIFLEFFAWGLLTAPTLVVLHETFPKHTFLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISPWWYFAVISVSGVFAVTFSVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAIGAYLGRVYGDSLVVVLATAIALLDICFILVAVPESLPEKMRPASWGAPISWEQADPFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLRQIMKFSPESVAAFIAVLGILSIIAQTIVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSITFPAVSALVSRTADADQQGVVQGMITGIRGLCNGLGPALYGFIFYIFHVELKELPITGTDLGTNTSPQHHFEQNSIIPGPPFLFGACSVLLALLVALFIPEHTNLSLRSSSWRKHCGSHSHPHSTQAPGEAKEPLLQDTNV
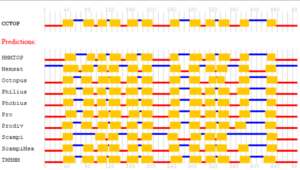
Supplemental information Figure 4: Predictions of membrane topology for the mouse mfsd14a protein. Obtained from the Constrained Consensus TOPology prediction server (http://cctop.enzim.ttk.mta.hu), where 10 sources/algorithms are combined to create a model. HMMTOP: Hidden Markov Model for Topology & Prediction; Memsat: MemSatSVM, Membrane helix prediction with support vector machines; Octopus: obtainer of correct topologies for uncharacterized sequences, a novel topology predictor (Viklund & Elofsson, 2008); Philius: a combined transmembrane topology and signal peptide predictor that extends Phobius by exploiting the power of dynamic Bayesian networks (DBN) (Reynolds et al., 2008); Phobius: A combined transmembrane topology and signal peptide predictor (Kall et al., 2004); Pro: Prodiv: profile-based hidden Markov model (Viklund & Elofsson, 2004); Scampi & ScampiMSA: first-principle-based topology predictors (Peters et al., 2016); TMHMM: Transmembrane Helices; Hidden Markov Model (protein topology), a tool used to predict the presence of transmembrane helices in proteins (Krogh et al., 2001).

Supplemental information Figure 5: NCBI protein report for Reference Sequence: NP_032272.2. Obtained on January 20th 2022, from
https://www.ncbi.nlm.nih.gov/protein/NP_032272.2?report=graph
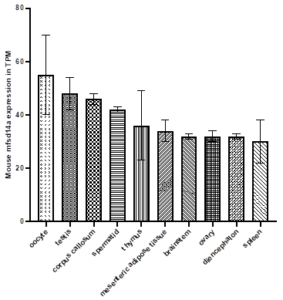
Supplemental information Figure 6: Top ten tissues for mfsd14a expression (displayed in Transcripts Per Million, TPM) in the adult mouse. Data Source: MGI mouse tissue expression database.

Supplemental information Figure 7: mfsd14a expression (displayed in Transcripts Per Million, TPM) in adult mouse tissues. The ten tissues of lowest expression. Data Source: MGI mouse tissue expression database.

Supplemental information Figure 8: mfsd14a expression in adult mouse brain from the Allen Mouse Brain Atlas (Allen Mouse Brain Atlas, http://mouse.brain-map.org/experiment/show/69734989)