Publication Information
ISSN: 2641-6816
Frequency: Continuous
Format: PDF and HTML
Versions: Online (Open Access)
Year first Published: 2018
Language: English
| Journal Menu |
| Editorial Board |
| Reviewer Board |
| Articles |
| Open Access |
| Special Issue Proposals |
| Guidelines for Authors |
| Guidelines for Editors |
| Guidelines for Reviewers |
| Membership |
| Fee and Guidelines |
 |
The Human Major Facilitator Superfamily Domain Containing 14A (MFSD14A) Gene Does Not Encode a Glucose Transporter
Zhouyao H1, Weihrauch D2 & Eck K P1*
1University of Manitoba, Department of Food and Human Nutritional Sciences, 66 Chancellors Cir, Winnipeg, MB R3T 2N2, Canada
2University of Manitoba, Department of Biological Sciences, 190 Dysart Road, Winnipeg, MB R3T 2N2, Canada
Received Date: February 07, 2022; Accepted Date: February 16, 2022; Published Date: February 22, 2022;
*Corresponding Author: Peter Karl Eck, University of Manitoba, Department of Food and Human Nutritional Sciences, 66 Chancellors Cir, Winnipeg, MB R3T 2N2, Canada. Email: Peter.Eck@umanitoba.ca
Citation: Zhouyao H, Weihrauch D & Eck K P (2022) The Human Major Facilitator Superfamily Domain Containing 14A (MFSD14A) Gene Does Not Encode a Glucose Transporter. Adv in Nutri and Food Sci: ANAFS-225.
DOI: 10.37722/ANAFS.2022102
Abstract
The literature record on the human MFSD14A gene is extremely limited. However, disease associations to infertility and cancer severity are emerging, which warrants a thorough interrogation of the genomic locus to establish a baseline annotation. It had been suggested that MFSD14A is a putative glucose solute carrier, however, this was never tested.
This report establishes the baseline on the human MFSD14A genomic locus, encoding one ubiquitously expressed transcript which translates into a protein of 490 amino acids. The protein is predicted to contain a binding pocket for glucose and similar monosaccharides, however, when expresses in Xenopus laevis oocytes doe not mediate uptake of radiolabeled 2-deoxy-glucose.
Introduction
To date, eight publications refer to the human MFSD14A gene (Supplemental information, Table 1). It is described to contain an “off target” target site in CRISPR/Cas9 intervention (Osborn et al., 2016), and a circular RNA derived from the human MFSD14A gene locus is implicated in the development of gastric (Quan et al., 2020) and renal cell carcinomas (Wang et al., 2017). In T-cell lymphoblastic lymphoma a novel fusion transcript containing SLC35A3-MFSD14A is identified (Lopez-Nieva et al., 2019). Significantly, no baseline data on the human MFSD14A genomic locus and its encoded products are in the publication record. This report aims to establish this.
Based on the analysis of the mouse protein, it is extrapolated that mouse mfsd14a might encode for a novel sugar transporter (Matsuo et al., 1997), however, no functional data are published. This report interrogates the glucose transport mediated by human MFSD14A. This might illuminate on the clinically relevance of the phenotype of male infertility resembling human globozoospermia in mfsd14a-/- mice (Doran et al., 2016).
Overall, in the light of emerging clinical relevance, this report aims to establish baseline data on the human MFSD14A genomic locus and interrogates its ability to mediate transmembrane transport of glucose.
Material and Methods
Bioinformatics genomics analysis
The genomic sequence of human MFSD14A (Chromosome 1 - NC_000001.11), full length RNA sequences including both reference and non-reference RNA sequences, and the expressed sequence tags (ESTs) sequences were retrieved from the National Center for Biotechnology Information (NCBI) database. Alignments between reference RNA sequence and the genomic DNA sequence was made in the DNA Sequence Analysis Software – Sequencher 4.8 (GeneCodes Corporation; Ann Arbor, Michigan, USA) to determine the structures of the exons and the introns (with large gap alignment parameter, 20 minimum overlap and 80% minimum matches). A scaled illustration was created after the exons and introns are manually curated to demonstrate the genomic organization and the structures of the exon and introns.
Protein analysis
The function and structure of the MFSD14A protein was analysed with the Protein Homology/analogY Recognition Engine V 2.0 (Phyre2, (Kelley et al., 2015)) and the AlphaFold Model using 3D ligand site (Jumper et al., 2021; Wass et al., 2010) with parameters from the standard/normal modes.
Expression patterns analysis
The following resources are used to obtain the experimental RNA-seq data on panels of adult human tissues: the European Molecular Biology Laboratory's European Bioinformatics Institute (EMBL-EBI) Baseline Expression Atlas (URL: https://www.ebi.ac.uk/gxa/home) (Moreno et al., 2022) and The Encyclopedia of DNA Elements (ENCODE) (URL: https://www.encodeproject.org, (Consortium, 2012). RNA-seq data from the following projects were interrogated: the Genotype-Tissue Expression (GTEx) project containing 53 samples (Consortium, 2015); The National Institutes of Health (NIH) Roadmap Epigenomics Mapping Consortium containing 19 samples (Roadmap Epigenomics et al., 2015), The Functional Annotation of the Mammalian Genome 5 (FANTOM5) project containing 56 samples (URL: https://fantom.gsc.riken.jp/data/), the Mammalian Kaessmann project containing 8 samples (Cardoso-Moreira et al., 2019) and the Encyclopedia of DNA Elements (ENCODE) project containing 13 samples (Lin et al., 2014). The RNA expression level of MFSD14A is presented in transcript per million (TPM).
Subcloning of MFSD14A
Plasmids containing the full open reading frames (ORF) of MFSD14A and GLUT3 (positive control for glucose transport study) was purchased from Harvard PlasmID (MFSD14A clone ID: HsCD00399291; GLUT3 clone ID: HsCD00021270) and transformed into DH5α competent cells (New England Biolabs, Ipswich, Massachusetts, USA) following the manufactures guidance for propagation, followed by column purification (QIAprep Spin Miniprep Kit, Qiagen, Hilden, Germany). The ORFs were then amplified with Q5 High Fidelity DNA Polymerase (New England Biolabs) and either tagged with restriction enzyme recognition sites (BamHI and XbaI) suitable for Xenopus laevis expression vector, PGEM-HE (for MFSD14A) or tagged with T7 RNA polymerase promoter sequence (for GLUT3) (primer sequences in Supplemental information Table 2). The plasmid containing human MFSD14A was then linearized by SphI prior to the in vitro transcription of capped mRNA (cRNA) (HiScribe T7 ARAC mRNA kit, New England Biolabs) whereas the GLUT3 amplicon containing GLUT3 ORF and T7 promoter sequence was used directly for cRNA synthesis as described previously. cRNA products were column purified (RNeasy MinElute Cleanup Kit, Qiagen) prior to the spectrophotometric quantification (Nanodrop, ND-1000, Thermofisher, Waltham, MA, USA) and the integrity assessments (MOPS agarose gel containing formaldehyde).
Oocyte experiments
Stage VI-V oocytes were collected from mature female Xenopus laevis (VWR International, Randor, PA, USA) as previously described ((Soreq & Seidman, 1992). Briefly, the frogs were euthanized via decapitation prior to the collection of the ovaries. The ovaries were placed in calcium-free OR2 solution (contained (in mmol l-1) 82.5 NaCl, 2.5 KCl, 1 MgCl2, 1 Na2HPO4, 5 HEPES, pH 7.4) with collagenase type VI (1 mg ml-1) (Gibco, Waltham, MA, USA), gently agitated for 90 minutes at room temperature. 1 mmol l-1 CaCl2 was added to terminate the collagenase activity. Oocytes were then manually sorted, rinsed three additional times with standard OR2 solution (contained (in mmol l-1) 82.5 NaCl, 2.5 KCl, 1 MgCl2, 1 Na2HPO4, 5 HEPES, 1 CaCl2, pH 7.4). For long term storage of isolated oocytes, the standard OR2 solution was supplemented with 2.5 mmol l-1 sodium pyruvate, 1 mg ml-1 penicillin-streptomycin (Gibco, Long Island, NY, USA) and 50 μg ml−1 gentamicin and held at 16 °C without the exposure of light. All procedures used were approved by the University of Manitoba Animal Research Ethics Board and are in accordance with the Guidelines of the Canadian Council on Animal Care.
cRNA injection
Isolated oocytes after the overnight recovery were injected with 18.4 ng of cRNA (36.8 nL with 0.5 ng nl-1) or nuclease-free water as negative control using the Nanoject II auto-nanoliter injector (Drummond Scientific, Broomall, Pennsylvania, USA). Experiments were conducted 48 hours post-injection.
Glucose uptake study in oocytes expressing MFSD14A and GLUT3
Oocytes expressing MFSD14A, GLUT3 (positive control) and SHAM (oocytes injected with water, negative control) were equilibrated at room temperature in standard OR2 solution two days post-injection, and randomly divided into groups containing 20 oocytes per group. Experiments were performed by incubating groups of oocytes in 200 μl standard OR2 solution which consisting of 125 pmol [3H] 2-deoxyglucose (25.5 Ci/mmol, PerkinElmer, Waltham, MA, USA) for 30 minutes. Excess ice-cold standard OR2 solution was added to terminate the transport, followed by four washes with the same solution. Oocytes were then individually lysed in scintillation vials (6 mL Pony Vial, PerkinElmer) with 200 μl of 10% sodium dodecyl sulfate (SDS) before the addition of 5 ml Ultima Gold scintillation cocktail (PerkinElmer). Internal radioactivity was quantified by liquid scintillation spectrometry as CPM oocyte-1 30min-1.
Results
The human MFSD14A genomic locus
The human MFSD14A gene is listed in NCBI as gene ID 64645, residing on the sense strand of human chromosome 1, more specific chr1:100,503,789-100,548,929 in 1p31 (GRCh37/hg19) (Figure 1). It is immediately flanked by the SAS-6 centriolar assembly protein (SASS6) and the solute carrier family 35 member A3 (SLC35A3) genes, which might explain the detected fusion protein (Lopez-Nieva et al., 2019).
The visually curated alignment of transcripts shows that the human MFSD14A gene spans 45,285 bases from its transcription initiation to the termination site. The transcript starts 155 nucleotides upstream of the site currently identified NCBI Reference Transcript NM_033055 (supplemental information Figure 1), while the transcriptional termination sites is correctly annotated.
The gene contains 12 exons transcribing into one transcript (NM_033055) (Figure 1), encoding Reference Protein NP_149044 (supplemental information Sequence 1).
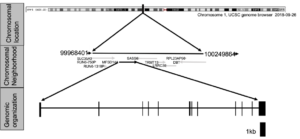
Figure 1: The human MFSD14A genomic locus and its encoded transcript.
The human MFSD14A protein
The human MFSD14A protein contains 490 amino acids, as represented by NCBI reference protein NP_149044 (supplemental information Sequence 1). Consistent with prior literature reports, the protein is predicted to be a solute membrane carrier spanning twelve transmembrane domains (Figure 2A and Supplemental information Figure 2).
The human MFSD14A protein clusters to the protein family MFS, subfamily MFSD14 (Supplemental information Figure 2C). This subfamily contains only one additional member, MFSD14B. Both family members contain a unique putative chemical substrate binding pocket, setting it somewhat aside from the glucose transporter families. However, the putative organic substrates, glucose, xylose, maltose, alanine, chloramphenicol, and 3ALPHA, 5BETA, 12ALPHA)-3, 12-DIHYDROXYCHOLAN-24-OIC ACID are predicted to bind in a pore like intramolecular binding pocket (Figure 2C), supporting others’ speculations that glucose might be a main substrate.
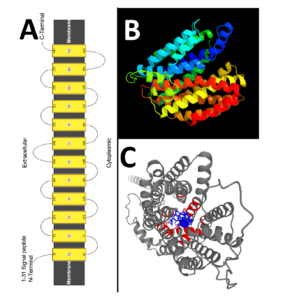
Figure 2: The predicted (panel A) secondary and (panel B) tertiary structures for the mouse mfsd14a protein (Image colored by rainbow N→C terminus; Model dimensions (Å):X:58.386 Y:50.944 Z:54.127). Models created using the phyre2 web-server. Panel D depicts the putative ligand binding pocked aligned in a “pore like” structure created as an AlphaFold Model using the 3D ligand site (https://www.wass-michaelislab.org/3dlig/web_results/61eae944c331a/61eae944c331a.html).
Tissues expression of the human MFSD14A transcript
Human MFSD14A expression is ubiquitous, no tissue interrogated is without evidence (Figure 3 and Supplemental information Figures 3A-D). Highest expression was observed in breast, spinal cord, seminal vesicles, skink testis (Figure 3), and renal areas (Supplemental information Figures 3A-D). In general, significant expression was exhibited by all tissues assessed, with lowest levels in muscle, liver, placenta, and thymus (Figure 3 and Supplemental information Figures 3A-D).
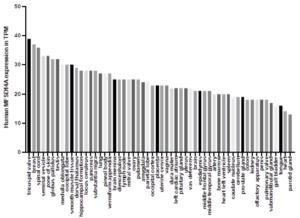
Figure 3: Human MFSD14A expression data from the FANTOM5 project. Data Source: EBI expression Atlas. Expression levels in transcripts per million, TPM.
Human MFSD4A does not mediate glucose uptake into Xenopus laevis oocytes.
When human MFSD14 is expressed in Xenopus laevis oocytes, no glucose uptake was detected (Figure 4). This provides evidence that glucose or similar saccharides are not a substrate.
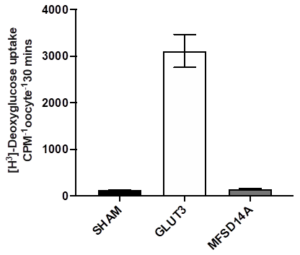
Figure4: The uptake of radiolabeled 2-deoxyglucose into Xenopus laevis oocytes expressing GLUT3 (known glucose carrier) and the human MFSD14A.
Discussion
This report establishes the basic annotation of the human MFSD14A locus, containing one transcript, and encoding one protein of 490 amino acids. This visually curated annotation corroborates the records in the NCBI, EMBL and UCSC genomic databases, except that the transcriptional start site is located 155 nucleobases upstream of the currently annotated. Tissue expression is ubiquitous with some of the highest levels detected in testis, which might indicate a relevance to the globozoospermia-like phenotype of the mfsd14a-/- mice (Doran et al., 2016). The baseline genomics established here should serve as a nucleus to interrogate the genetic of this very rare human condition (Modarres et al., 2019).
Evolutionary conservation of MFSD14A is high (Doran et al., 2016; Lekholm et al., 2017), which is confirmed by the presence of a sole transcript and protein, since alternative splicing seems to negatively correlate with fitness of the locus (Saudemont et al., 2017). This also indicates a basic ubiquitous function, and it could be expected that more than fertility might be affected in gene disruption. More mechanistic insight will be possible upon the identification of the MFSD14A substrate.
The human MFSD14A does not mediate transmembrane glucose uptake. This dismisses the strong suggestion voiced in prior report (Matsuo et al., 1997) and indicated by here presented bioinformatics prediction, that glucose could be a canonical substrate of the MFSD14A protein. Although this does not rule out other monosaccharides as MFSD14A substrates, it limits the substrate range to fructose/fructose like molecules, since the multi-specific substrate recognition reported for the SLC2A (GLUT) and SLC5A (SGLT) transporters family members include glucose, xylose, arabinose, and mannose (Deng & Yan, 2016). This severely limits the likelihood of any of these substrates been recognized by MFSD14A.
In the quest to deorphanize the MFSD14A gene/protein, data from other species might give novel leads: the mouse mfsd14a is differentially regulated in response to amino acid and food deprivation, as well as high fat diet (Lekholm et al., 2017). Although this compels a strong connection to energy/carbohydrate regulation, it could also point towards a function in amino acids or lipids pathways. Moreover, mfsd14a was differentially regulated in the gills of the crab Carcinus maenas acclimated to brackish water (Fehsenfeld et al., 2011). Specifically, after challenges with high pCO2 and ammonia, mfsd14a was downregulated (Fehsenfeld et al., 2011). This indicates a potential involvement in detoxification/acid-base pathways, with ammonium as a prime candidate.
Ubiquitous expression in relevant levels could link MFSD14A to cancers beyond gastric cancer and renal cell carcinomas (Li et al., 2017; Quan et al., 2020; Wang et al., 2017), since the expression of the disease modulating circular RNA circHIAT1 is correlated with the general expression of the gene.Therefore, this repot should establish the baseline expression to inform future research avenues.
Conclusion
The ubiquitously expressed human MFSD14A gene transcribes into a single transcript encoding one protein. It does not mediate glucose uptake into Xenopus laevis oocytes. The genomic and functional baseline to inform on human diseases is established.
Reference
- Cardoso-Moreira M, Halbert J, Valloton D, Velten B, Chen C, et al. (2019). Gene expression across mammalian organ development. Nature, 571:505-509.
- Consortium E P (2012). An integrated encyclopedia of DNA elements in the human genome. Nature, 489:57-74.
- Consortium G T (2015). Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348:648-660.
- Deng D & Yan N (2016). GLUT, SGLT, and SWEET: Structural and mechanistic investigations of the glucose transporters. Protein Sci, 25:546-558.
- Doran J, Walters C, Kyle V, Wooding P, Hammett-Burke R, et al. (2016). Mfsd14a (Hiat1) gene disruption causes globozoospermia and infertility in male mice. Reproduction, 152:91-99.
- Fehsenfeld S, Kiko R, Appelhans Y, Towle D W, Zimmer M, et al. (2011). Effects of elevated seawater pCO(2) on gene expression patterns in the gills of the green crab, Carcinus maenas. BMC Genomics, 12, 488.
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature, 596:583-589.
- Kelley L A, Mezulis S, Yates C M, Wass M N, Sternberg M J (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc, 10:845-858.
- Lekholm E, Perland E, Eriksson M M, Hellsten S V, Lindberg F A, et al. (2017). Putative Membrane-Bound Transporters MFSD14A and MFSD14B Are Neuronal and Affected by Nutrient Availability. Front Mol Neurosci, 10, 11.
- Li P, Chen H, Chen S, Mo X, Li T, et al. (2017). Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer, 116:626-633.
- Lin S, Lin Y, Nery J R, Urich M A, Breschi A, et al. (2014). Comparison of the transcriptional landscapes between human and mouse tissues. Proc Natl Acad Sci USA, 111:17224-17229.
- Lopez-Nieva P, Fernandez-Navarro P, Grana-Castro O, Andres-Leon E, Santos J, et al. (2019). Detection of novel fusion-transcripts by RNA-Seq in T-cell lymphoblastic lymphoma. Sci Rep, 9:5179.
- Matsuo N, Kawamoto S, Matsubara K, Okubo K (1997). Cloning of a cDNA encoding a novel sugar transporter expressed in the neonatal mouse hippocampus. Biochemical and Biophysical Research Communications, 238: 126-129.
- Modarres P, Tavalaee M, Ghaedi K, Nasr-Esfahani M H (2019). An Overview of the Globozoospermia as a Multigenic Identified Syndrome. Int J Fertil Steril, 12:273-277.
- Moreno P, Fexova S, George N, Manning J R, Miao Z, et al. (2022). Expression Atlas update: gene and protein expression in multiple species. Nucleic Acids Res, 50:D129-D140.
- Osborn M J, Webber B R, Knipping F, Lonetree C L, Tennis N, et al. (2016). Evaluation of TCR Gene Editing Achieved by TALENs, CRISPR/Cas9, and megaTAL Nucleases. Mol Ther, 24:570-581.
- Quan J, Dong D, Lun Y, Sun B, Sun H, et al. (2020). Circular RNA circHIAT1 inhibits proliferation and epithelial-mesenchymal transition of gastric cancer cell lines through downregulation of miR-21. J Biochem Mol Toxicol, 34:e22458.
- Roadmap Epigenomics C, Kundaje A, Meuleman W, Ernst J, Bilenky M, et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature, 518:317-330.
- Saudemont B, Popa A, Parmley J L, Rocher V, Blugeon C, et al. (2017). The fitness cost of mis-splicing is the main determinant of alternative splicing patterns. Genome Biol, 18:208.
- Wang K, Sun Y, Tao W, Fei X, Chang C (2017). Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett, 394:1-12.
- Wass M N, Kelley L A, Sternberg, M J (2010). 3DLigandSite: predicting ligand-binding sites using similar structures. Nucleic Acids Res, 38(Web Server issue), 38:W469-473.
Supplement Information:
Supplemental Information Table 1: List of currently available published literature on MFSD14A
Authors
Title
Year
Journal
Quan, J., Dong, D., Lun, Y., Sun, B., Sun, H., Wang, Q., & Yuan, G.
Circular RNA circHIAT1 inhibits proliferation and epithelial-mesenchymal transition of gastric cancer cell lines through downregulation of miR-21.
2020
The Journal of Biochemical and Molecular Toxicology
Lekholm, E., Perland, E., Eriksson, M. M., Hellsten, S. V., Lindberg, F. A., Rostami, J., & Fredriksson, R.
Putative Membrane-Bound Transporters MFSD14A and MFSD14B Are Neuronal and Affected by Nutrient Availability.
2017
Frontiers in Molecular Neuroscience
Li, P., Chen, H., Chen, S., Mo, X., Li, T., Xiao, B., Yu, R., Guo, J.
Circular RNA 0000096 affects cell growth and migration in gastric cancer
2017
British Journal of Cancer
Wang, K., Sun, Y., Tao, W., Fei, X., & Chang, C.
Androgen receptor (AR) promotes clear cell renal cell carcinoma (ccRCC) migration and invasion via altering the circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals.
2017
Cancer Letters
Doran, J., Walters, C., Kyle, V., Wooding, P., Hammett-Burke, R., & Colledge, W. H.
Mfsd14a (Hiat1) gene disruption causes globozoospermia and infertility in male mice
2016
Reproduction
Osborn, M. J., Webber, B. R., Knipping, F., Lonetree, C. L., Tennis, N., DeFeo, A. P., . . . Blazar, B. R.
Evaluation of TCR Gene Editing Achieved by TALENs, CRISPR/Cas9, and megaTAL Nucleases.
2016
Molecular Therapy
Fehsenfeld, S., Kiko, R., Appelhans, Y., Towle, D. W., Zimmer, M., & Melzner, F.
Effects of elevated seawater pCO(2) on gene expression patterns in the gills of the green crab, Carcinus maenas.
2011
BMC Genomics
Matsuo, N., Kawamoto, S., Matsubara, K., & Okubo, K.
Cloning of a cDNA encoding a novel sugar transporter expressed in the neonatal mouse hippocampus
1997
Biochemical and Biophysical Research Communication
Name
Sequence 5' -> 3'
GLUT3 ORF forward
ATGGGGACACAGAAGGTCACCCCAG
GLUT3 ORF reverse
TTAGACATTGGTGGTGGTCTCCTTA
GLUT3 T7 forward
AAAATAATACGACTCACTATAGACCATGGGGACACAGAAGGTCAC
MFSD14A ORF forward
ATGACCCAGGGGAAGAAGAAGAAAC
MFSD14A ORF reverse
TCACACATTTGTGTCCTGGAGTAAA
MFSD14A BamHI forward
AAAAGGATCCACCATGACCCAGGGGAAGAAGAAGAAAC
MFSD14A XbaI reverse
AAAATCTAGATCACACATTTGTGTCCTGGAGTAAA
Supplemental information Sequence 1: the amino acid sequence of the human MFSD14A protein.
>NP_149044.2 MFSD14A [Homo sapiens]
MTQGKKKKRAANRSIMLAKKIIIKDGGTPQGIGSPSVYHAVIVIFLEFFAWGLLTAPTLVVLHETFPKHT
FLMNGLIQGVKGLLSFLSAPLIGALSDVWGRKSFLLLTVFFTCAPIPLMKISPWWYFAVISVSGVFAVTF
SVVFAYVADITQEHERSMAYGLVSATFAASLVTSPAIGAYLGRVYGDSLVVVLATAIALLDICFILVAVP
ESLPEKMRPASWGAPISWEQADPFASLKKVGQDSIVLLICITVFLSYLPEAGQYSSFFLYLRQIMKFSPE
SVAAFIAVLGILSIIAQTIVLSLLMRSIGNKNTILLGLGFQILQLAWYGFGSEPWMMWAAGAVAAMSSIT
FPAVSALVSRTADADQQGVVQGMITGIRGLCNGLGPALYGFIFYIFHVELKELPITGTDLGTNTSPQHHF
EQNSIIPGPPFLFGACSVLLALLVALFIPEHTNLSLRSSSWRKHCGSHSHPHNTQAPGEAKEPLLQDTNV
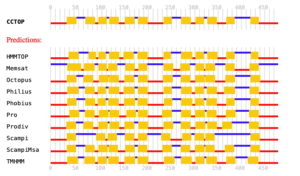
Supplemental information Figure 2: Predictions of membrane topology for the mouse mfsd14a protein. Obtained from the Constrained Consensus TOPology prediction server (http://cctop.enzim.ttk.mta.hu), where 10 sources/algorithms are combined to create a model.
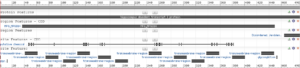 Supplemental information Figure 2: NCBI protein report for Reference Sequence: NP_149044.2. Obtained on January 22nd 2022, from https://www.ncbi.nlm.nih.gov/protein/NP_149044.2?report=graph
Supplemental information Figure 2: NCBI protein report for Reference Sequence: NP_149044.2. Obtained on January 22nd 2022, from https://www.ncbi.nlm.nih.gov/protein/NP_149044.2?report=graph

Supplemental information Figure 3A: Expression of MFSD14A in adult human tissues. Data Source: EBI expression Atlas the GTEx project. Expression levels in transcripts per million, TPM.
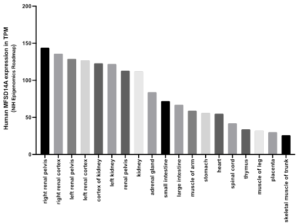
Supplemental information Figure 3B: Expression of MFSD14A in adult human tissues. Data Source: EBI expression Atlas the NIH Epigenomics Roadmap project. Expression levels in transcripts per million, TPM.

Supplemental information Figure 3C: Expression of MFSD14A in adult human tissues. Data Source: EBI expression Atlas the Kaessman project. Expression levels in transcripts per million, TPM.

Supplemental information Figure 3D: Expression of MFSD14A in adult human tissues. Data Source: EBI expression Atlas the ENCODE project. Expression levels in transcripts per million, TPM.