Publication Information
Frequency: Continuous
Format: PDF and HTML
Versions: Online (Open Access)
Year first Published: 2019
Language: English
| Journal Menu |
| Editorial Board |
| Reviewer Board |
| Articles |
| Open Access |
| Special Issue Proposals |
| Guidelines for Authors |
| Guidelines for Editors |
| Guidelines for Reviewers |
| Membership |
| Fee and Guidelines |
 |
Molecular Analysis of the Genetic Relationship and the Attribution of Some Unsubstantiated Resources of Hawthorn (Crataegus)
LiyanYu1, 2, ZhiheWang2, Xiao Du1, Xiao Zhang 1, Wenxuan Dong1*
1College of Horticulture, Shenyang Agricultural University, Shenyang, Liaoning 110866, China
2Weifang University of Science and Technology, Shouguang 262700, China
Received Date: August 08, 2021; Accepted Date: August 30, 2021; Published Date: September 11, 2021
*Corresponding author: Wenxuan Dong, College of Horticulture, Shenyang Agricultural University, 120, Dongling Road, Shenhe District, Shenyang City, Liaoning Province, 110866, China. Tel: +8613898813246; Email: dongwx63@syau.edu.cn
Citation: Yu L, Wang Z, Du X, Zhang X, Dong W (2021) Molecular Analysis of the Genetic Relationship and the Attribution of Some Unsubstantiated Resources of Hawthorn (Crataegus). Adv Agri Horti and Ento: AAHE-159.
DOI: 10.37722/AAHAE.2021602
Abstract
The genus Crataegus (hawthorn), belonging to the subfamily Maloideae in the Rosaceae family, has a long history of cultivation in China because of its important medicinal values. we carried out a phylogenetic reconstruction with nuclear (ITS) and three intergenic chloroplast DNA sequences (trnH-trnK, trnG-trnS, and psbA-trnH) data using ML and MP to estimate the genetic relationships, chloroplast haplotypes, and the origin of 17 species and 108 accessions of Crataegus and the attribution of some unsubstantiated resources in China. Malus baccata L. and Malus pumila L. were used as outgroups taxa. We further analyzed the basic phylogenetic framework of the genus. Our analyses produced multiple outcomes: (1) Crataegus in northern and southern China were divided into two branches, which had their respective origin relations and different speciation events. Crataegus might have originated in Europe and North America; (2) the classification of most samples based on the molecular data was in good agreement with the morphological classification. However, incongruence between the chloroplast and nuclear data supported the hypothesis of a hybrid origin for Crataegus brettschneideri Schneid, with Crataegus maximowiczii Schneid or its ancestor as the maternal parent and Crataegus pinnatifida Bunge as the male parent; (3) Chloroplast haplotypes and haplotype network graph analyses revealed 15 haplotypes among the specimens. H12 (Crataegus cuneata Sieb.) was a chloroplast ancestral haplotype of Crataegus in southern China, and H14 might be the direct origin haploid of the Pinnatifidae group.
Keywords: Chloroplast Haplotypes; Crataegus; Genetic Relationship; Hawthorn Resources; ITS; Molecular Evolution
Introduction
The genus Crataegus L., a common deciduous fruit tree commonly known as hawthorn, belongs to the subfamily Maloideae in the Rosaceae family. This genus has a long history of cultivation with a rapidly developing cultivation area in China, especially during the last century, because their roots, stems, leaves, and especially fruits, have important economic value in traditional Chinese medicine. Hawthorn has therapeutic benefits owing to its unique medicinal ingredients, including biologically active compounds such as phenols, flavonoids, and oligomeric procyanidins [1]. Laboratory tests and clinical trials have shown that hawthorns can be used for the treatment and prevention of cardiovascular diseases [2]. All species of this genus are shrubs to small trees. They are distributed widely in northern temperate regions, especially in Eurasia and North America, and approximately 140–200 species throughout the northern hemisphere have been described [3]. The earliest fossils have shown that this genus dates from the mid-Tertiary [4]. China is both the origin and the center of hawthorn cultivation [3]. Previous studies, based on geographical localities and morphologies, have suggested that there are 18 species and 6 varieties of Crataegus that are widely distributed in [5, 6]. A more recent study has shown that there are 20 species and 7 varieties of the Chinese Crataegus [7]. Phipps [3, 8] suggested that there were two basal species in the genus Crataegus based on cladistic analyses of morphological data; one from southern China and the other from Mexico. This author further postulated that the trans-Beringian migration of Asian and American Crataegus resulted in the modern distribution of the genus. The migration may have occurred via two paths; one westward from southwest China to Europe and the other eastward from Eastern Asia to North America. He suggested that Crataegus scabrifolia Rehd. Evolved into the European Crataegus and other Chinese Crataegus species (Crataegus pinnatifida Bunge, Crataegus hupehensis Sarg. and Crataegus sanguinea Pall.) [3]. However, this hypothesis was later contradicted by that of the North American origin [9]. New evidence from a study of sequences of the internal transcribed spacer (ITS) region, chloroplast DNA (cpDNA) regions, and LEAFY intron2 has suggested that Crataegus originated in eastern North America and Europe [10]. Zarrei et al. [11] studied sequence data from 14 plastid loci and suggested the origin of the section Sanguineae (Crataegus maximowiczii Schneidas the section) involved the east-to-west trans-Beringian migration from western North America into eastern Asia.
Molecular markers were used to determine genetic relationships within plant populations, and the reliability were almost 100% [12]. Simple sequence repeats (SSRs) [10, 13, 14], random amplified repeats [15, 16], and inter-simple sequence repeats (ISSRs) [17, 18] have been used for genetic characterization of Crataegus and analysis of genetic diversity of Crataegus. Intraspecific [17, 19-22] and interspecific [23-25] relationships of Chinese Crataegus have also been studied. Du et al. [25] explored the origin and evolution of 53 cultivated Crataegus and three related species that were native to China at the genomic level based on SSRs and single-nucleotide polymorphisms.
Previous studies provided useful information that partially resolved the phylogenetic history of and relationship among Crataegus species in China. However, the interspecific relationship and evolution of hawthorns, as well as the genetic relationship and attribution of some unsubstantiated resources of hawthorn (Crataegus) based on chloroplast simple sequence repeats (cpSSRs) [26] and ribosomal ITS in China remain unclear. In the present study, we attempted to determine the genetic relationships among 17 species and 108 accessions of Crataegus and the attribution of some unsubstantiated resources of Crataegus based on ribosomal ITS and three intergenic cpDNA regions.
Materials and Methods
Tax on Sampling
Species
No. of individuals
Regions
C. pinnatifida Bunge
10, 1
China, Korea
C. pinnatifida Bge.var. major N.E.Br.
47
China
C. brettschneideri Schneid
3
China
C. jozana Schneid
1
Japan
C. hupehensis Sarg.
5
China
C. scabrifolia Rehd.
3
China
C. songarica Koch
1
China
C. cuneata Sieb.et Zucc
16
China
C. altaica (Loud.) Lange.
3
China
C. dahurica Schneid
1
China
C. kansuensis Wils.
1
China
C. maximowiczii Schneid
2, 1
China, Russia
C. sanguinea Pall.
1, 3
China, Russia
C. chlorosarca Maxim.
1
Russia
C. laevigata Poir.
1
Britain
Mespilus germanica L.
1
Czech Republic
C. monogyna Jacq.
1
Russia
HSD
2
North America
J6
1
Canada
ZWSLH
1
China
GSSZ
1
China
In this investigation, 108 accessions (Table 1) and three out groups were sampled. They were broadly distributed across a range of geographical and climatic conditions and thus were representative of Chinese hawthorn diversity and encompassed all possible introductory sources in China. The samples were selected as representatives of Chinese Crataegus in previous phylogenetic studies [3, 10, 11, 25, 27], including four species cultivated in China (C. hupehensis, C. pinnatifida var.major, C. brettschneideri, and C. scabrifolia), eight species distributed close to the cultivated Crataegus, which are widely distributed and cover most of the different regions in China. C. scabrifoliais considered to be the ancestral Crataegus species. C. pinnatifida is a species that is widespread throughout China. C. pinnatifida var. major endemic to China and has the longest cultivation history. Accession species of Malus pumila Mill. and Malus baccata L. were used as outgroups. Samples were either collected in the field or obtained from the National Hawthorn Germplasm Repository at Shenyang Agricultural University, China (Table 2). All materials tested were identified based on recent floristic and taxonomic references, such as the treatment of Crataegus in the Flora of China [28] and China fruit plant monograph of Hawthorn (Crataegus) flora [5].
Group
Taxon
ID
Biogeographic regions
Group
Taxon
ID
Biogeographic regions
Pinnatifidae
C. pinnatifida
JSTSLH
Heilongjiang,China
C. brettschneideri
FLH
Liaoning, China
SHSLH
Heilongjiang,China
ZF1H
Jilin, China
SZ
Liaoning, China
JF1H
Jilin, China
HGSLH
Korea
C. jozana
MHSlH
Japan
XKSLH
Heilongjiang,China
MDFSLH
Heilongjiang,China
Henryanae
C. hupehensis
JT
Hubei, China
SLH1
Liaoning, China
HBSZt
Zhejiang, China
SLH1
Liaoning, China
MHL
Shandong, China
SLH3
Liaoning, China
XP1H
Shandong, China
SLHBL
Liaoning, China
TASSZM
Shandong, China
ZRDZ
Liaoning, China
C. scabrifolia
YNSZ1H
Yunnan, China
C. pinnatifida Var.Major
HLH
Beijing,China
YNSZ2H
Yunnan, China
JD1H
Beijing,China
YNSZw
Yunnan, China
NJY2H
Beijing,China
Cuneatae
C. cuneata
AG1
Anhui, China
XZS4H
Beijing,China
AG2
Anhui, China
HBY1H
Hebei,China
AG3
Anhui, China
XLSS
Hebei,China
AG4
Anhui, China
XLZR
Hebei,China
ZS1
Zhejiang, China
YRQ
Hebei,China
ZS2
Zhejiang, China
ZH153H
Hebei,China
ZS3
Zhejiang, China
BQ780
Henan, China
ZS4
Zhejiang, China
LXSK
Henan, China
ZS5
Zhejiang, China
YBH
Henan,China
ZD1
Zhejiang, China
GY2H
Jiangsu, China
ZD2
Zhejiang, China
XZDH
Jiangsu,China
ZD3
Zhejiang, China
JAZR
Jilin, China
ZD4
Zhejiang, China
JLYH
Jilin, China
YSZw
Henan, China
ASZR
Liaoning, China
YSZ2
Henan, China
DLQK
Liaoning, China
YSZ3
Henan, China
FS
Liaoning, China
Sanguineae
C. altaica
AET1
Xinjiang, China
XBRZ
Liaoning, China
AET2
Xinjiang, China
GDSZ1
Liaoning, China
AET3
Xinjiang, China
GDSZ2
Liaoning, China
C. maximowiczii
MSZ
Heilongjiang, China
GDSZ3
Liaoning, China
NASZ
Heilongjiang, China
JH
Liaoning, China
S4
Russia
LH
Liaoning, China
C. chlorosarca
HGLR
Russia
LYZR
Liaoning, China
C. sanguinea
S1
Russia
MP
Liaoning, China
S6
Russia
QJX
Liaoning, China
S7
Russia
TS
Liaoning, China
LNSZ
Jilin, China
XFH
Liaoning, China
C. dahurica
GYSZ
Liaoning, China
XH
Liaoning, China
C. kansuensis
GSSZ
Shanxi, China
XFRZ
Liaoning, China
Orientales
C. songarica
ZGE
Xinjiang, China
RR2
Liaoning, China
Laevigata
C. laevigata
HHSZ
Britain
RR4
Liaoning, China
Mespilus
M. germanica
germanica
Czech Republic
YR4H
Liaoning, China
C. monogyna
DZSZ
Russia
BRM
Shandong, China
Unknown
J6
Canada
FSMQ
Shandong, China
Unknown
HSD
North America
FSTQ
Shandong, China
Unknown
HSDw
North America
FXMQ
Shandong, China
Unknown
ZWSLH
Liaoning, China
HGSZ
Shandong, China
Unknown
GSSZ
Henan, China
SLZR
Shandong, China
XHM
Shandong, China
Malus baccata
SDZ
Liaoning, China
YDCK
Shandong, China
Malus pumila
HF
Liaoning, China
YDXH
Shandong, China
LYBNS
Shandong, China
JXSZ
Shanxi, China
LH1H
Shanxi, China
DNA Extraction, PCR Amplification and Sequencing
All samples collected were young leaves. Some were first collected in the field with an ice-box or liquid nitrogen, and then returned to the laboratory and stored at −80℃ until DNA extraction. Others were dried in the field with silica gel after collection and then stored in new silica gel until subsequent DNA extraction. We extracted total DNA from the leaves using a small-scale modified method of cetyl-trimethyl ammonium bromide according to the protocol described by Doyle [29]. The DNA quality was checked using a Nanodrop-2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Chloroplast primer sequences and ribosome ITS (ITS1-5.8 S-ITS2 region) markers used in the study are shown in Table 3. These primers were designed based on previous studies. PCR products were sequenced by direct sequencing and all samples were sequenced by GENEWIZ Biotechnology Co., Ltd. (Beijing, China). A total of 108 samples were analyzed.
Name
Forward primer sequences
Reverse primer sequences
psbA-trnH
GTTATGCATGAACGTAATGCTC
CGCGCATGGTGGATTCACAATCC
trnG- trnS
GAACGAATCACACTTTTACCAC
GCCGCTTTAGTCCACTCAGC
trnH-trnk
ACGGGAATTGAACCCGCGCA
CCGACTAGTTCCGGGTTCGA
ITS4
GGAAGGAGAAGTCGTAACAAGG
TCCTCCGCTTATTGATATGC
A PCR amplification reaction mixture with a total volume of 20 μL was prepared with 1.5 µL template DNA (20–30 ng), 9 µl of 2x Taq Master Mix buffer (GENEWIZ Biotechnology), 0.8 µL primers (20 ng/µl), and 8.7 µL sterile nuclease-free distilled H2O. The PCR protocol was as follows: initial denaturation at 94 ℃ for 4 min; 35 cycles of a three-step phase consisting of incubations at 94 ℃ for 45 s, 58 ℃ for 45 s, and 72 ℃ for 2 min; and a final extension step at 72 ℃ for 10 min. After the amplification was completed, the samples were stored at 4 ℃ [30]. PCR amplification was performed in a Thermal Cycler (Applied Biosystems, Foster City, CA, USA). The PCR products were sent to GENEWIZ Biotechnology Co., Ltd. for purification and sequencing.
Analytical Method of Sequence Alignment and Phylogenetic
Sequences of the examined regions were aligned separately using the software Geneious (Drummond et al. 2009) and manually adjusted. Gaps that were parsimony informative were coded into multistate characters using Seq State version 1.32 [31] and appended to the sequence matrices. Pairwise divergences among taxa for chloroplast and nuclear regions were estimated using the DNA maximum likelihood (ML) program of phylogeny inference package (PHYLIP) version 3.63 (Felsenstein 2006).
There were two main reasons for using different datasets to construct the phylogenetic trees. The first was to infer relationships among species from major biogeographical areas in China and the second was to further resolve the relationship between the cultivated and wild species of hawthorns. These were achieved by single and combined analyses of three cpDNA regions and one ITS region.
All datasets were analyzed using the maximum parsimony (MP) with equally weighted characters and maximum likelihood (ML) approaches in PAUP 4.0a165 [32] and PHYLIP version 3.63 (Felsenstein 2006), and DNAML, CONSENSE, and neighbor-joining (NJ) in MEGA. We inferred admixture graphs using Figtree v.1.4.3 [33].
The MP tree was randomly sampled by PAUP 4.0a165 software [32]. A total of 1000 samples were taken and divided into four parallel groups. Finally, the sample trees that were obtained four times were summarized to calculate the most consistent trees, and the bootstrapping results were annotated on each branch of the MP tree obtained previously by Adobe Illustrator CS6.The chloroplast haplotype was analyzed by DnaSP, the chloroplast haplotype network structure was analyzed by TCS, and the phylogenetic tree of the haplotype was analyzed by NJ using MEGA and Geneious.
Results
Analysis of cpDNA Cluster
From 18 pairs of spare primers for chloroplast gene fragments, we finally selected three pairs of primers with the largest number of mutation sites, including trnH-trnK, psbA-trnH, and trnG-trnS, for the amplification and sequencing of cpDNA. The molecular tree was constructed from these three pairs of chloroplast gene sequences (Figure 1). Using M. baccata and M. pumila as outgroups, Sanguineae group members, including C. altaica, C. maximowiczii, Crataegus chlorosarca, C. sanguinea, Crataegus dahurica, and Crataegus kansuensis, C. brettschneideri, and unsubstantiated materials ZWSLH and GSSZ formed a large clade NC, which was first separated to form a sister line with the other samples. Then, J6 from Canada, HSD from the United States formed NAclade. Mespilus germanica, Crataegus monogyna, and Crataegus laevigata from Europe, and Crataegus songarica from Xinjiang Province formed EU clade.
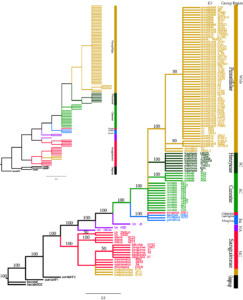
Figure 1: Chloroplast molecular phylogenetic tree of the 108 Crataegus samples.
(Notes: Strict consensus tree from maximum parsimony (MP) analyses using PAUP 4.0a165 software based on the combined trnH-trnK, psbA-trnH, and trnG-trnS chloroplast data. The inset (upper left corner) shows maximum likelihood (ML) using PHYLIP version 3.63 to illustrate branch lengths (drawn proportionally to the amount of change). Nodes with bootstrap values >50% are indicated. Accession species of Malus pumila Mill. and Malus baccata L. were used as outgroups. Bars indicate the biogeographic distribution of the examined taxa. NA, North America; NC, North China; SC, South China; and EU, Europe. Group, Region, and ID of examined individuals are found in Table 2.)
All remaining samples (including C. pinnatifida, Crataegus cuneata, C. hupehensis, C. scabrifoliaand soft-fleshed RR and hard-fleshed materials YR) formed SC clade. The first group to separate was the Cuneatae group, followed by the Henryanae group (C. hupehensis and C. scabrifolia), and then the Pinnatifidae group. These three groups formed a sisterhood. In the Pinnatifidae group, C. pinnatifida HGSLH from South Korea and C. pinnatifida SZ from NHZR separated first. Then, C. pinnatifida JSTSLH and C. pinnatifida SHSLH from Heilongjiang Province, and C. pinnatifida XBRZ from Xinbin of Liaoning Province separated. The remaining samples of C. pinnatifida, RR, YR, Crataegus jozana from Japan were completely clustered together.
Analysis of ITS DNA Cluster
A molecular tree was constructed based on the sequencing results of the amplification of all samples by ITS sequence (ITS1-5.8 S-ITS2 region) primers, and the results are shown in Figure 2. All materials were divided into two major clades. The first clade included NC, NA, and EU. The second clade contained only SC. Unlike in the chloroplast molecular tree, EU clade was isolated first, followed by the NA clade, and it was the sister to the Sanguineae group; Cuneatae group (C. cuneata) was the sister to Pinnatifidae group and Henryanae group. Hawthorn samples from southern and northern China were separated into two large clades, which revealed that these hawthorns had a distant genetic relationship and different origins.
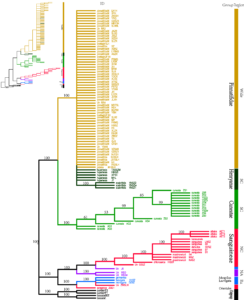
Figure 2: ITS molecular phylogenetic tree of the 108 Crataegus samples.
(Notes: Strict consensus tree from maximum parsimony (MP) analyses using PAUP 4.0a165 software based on ITS4 data. The inset (upper left corner) shows maximum likelihood (ML) using PHYLIP version 3.63. Nodes with bootstrap values >50% are indicated. Accession species of Malus pumila Mill. and Malus baccata L. were used as outgroups. Bars indicate the biogeographic distribution of the examined taxa. NA, North America; NC, North China; SC, South China and EU, Europe. Group, Region, and ID of examined individuals are found in Table 2.)
Analysis of Total DNA Cluster Analysis
A molecular phylogenetic tree was constructed based on the sequencing results of the amplification of all samples by three pairs of cpDNA sequences and ITS sequence (ITS1-5.8 S-ITS2) primers (Figure 3).
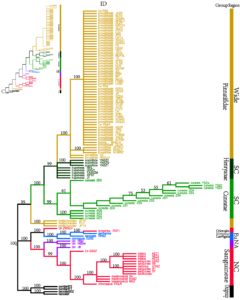
Figure 3: cp-ITS molecular phylogenetic tree of the 108 Crataegus.
(Notes: Strict consensus tree from maximum parsimony (MP) analyses using PAUP 4.0a165 software basing on the combined trnH-trnK, psbA-trnH, and trnG-trn Schloroplast data and ITS (ITS1-5.8 S-ITS2 region) data. The inset (upper left corner) shows maximum likelihood (ML) using PHYLIP version 3.63. Nodes with bootstrap values > 50% are indicated. Accession species of Malus pumila Mill. and Malusbaccata L. were used as outgroups. Bars indicate the biogeographic distribution of the examined taxa. NA, North America; NC, North China; SC, South China and EU, Europe. Group, Region and ID of examined individuals are found in Table 2.)
All tested samples were divided into two large clades, and their relationship was consistent with the molecular tree based on ITS alone. The difference was that C. brettschneideri separated from C. pinnatifida and formed a separate lineage, and C. hupehensis and C. scabrifolia separated from C. pinnatifida to form a separate lineage and was at the base of C. pinnatifidaclade. C. cuneata was divided into two sister lineages by region, indicating segregation between regions.
The results of the MP tree were consistent with those of the ML and NJ trees. Crataegus in northern China comprising C. altaica, C. maximowiczii, C. chlorosarca, C. sanguinea, C. dahurica, C. kansuensis, and GSSZ were part of the Sanguineae group. American hawthorn species J6 from Canada and HSD from the United States constituted a sisterhood with the Sanguineae group, which indicated that they were more closely related to hawthorn species in northern China. C. songarica was part of the Orientales group and shared a clade with M. germanica from Europe, C. laevigata from Britain, C. monogyna from Russia, and ZWSLH. Overall, the classification of some samples based on the molecular data was inconsistent with the morphological classification.
Analysis of Chloroplast Haplotypes
A total of 15 haplotypes were formed from 108 samples based on DnaSP (Table 4, Figure 4). The Sanguineae group from northern China was composed of C. maximowiczii, C. altaica, C. chlorosarca, and C. brettschneideri (H1); C. sanguinea, C. dahurica, and C. kansuensis (H3); and ZWSLH from Zhangwu of Liaoning Province and GSSZ from Guanshan of Henan Province (H4). C. altaica from Xinjiang Province had the unique haplotype H2, yet the only representative of the Orientales group, C. songarica, from Xinjiang Province had H8, and M. germanica, C. monogyna, and C. laevigata from Europe had H7. J6 from Canada had haplotype H5 and HSD from America H6. Henryanae group members from southeastern China, C. hupehensis and C. scabrifolia, shared haplotype H10, and the multispecies C. hupehensis JT had the unique haplotype H9. The Cuneatae group member C. Cuneata from Anhui and Zhejiang Provinces exhibited two haplotypes H11 and H12. The Pinnatifidae group member C. pinnatifida belonged to different haplotypes of H13, H14, and H15. C. pinnatifida SZ from NHZR and C. pinnatifida HGSLH from Korea belonged to the same haplotype H13, C. pinnatifida JSTSLH and C. pinnatifida SHSLH from Heilongjiang Province and C. pinnatifida XBRZ from Xinbin of Liaoning Province shared the haplotype H14, and others of C. pinnatifida (including C. jozana from Japan) belonged to the same haplotype H15. C. brettschneideri Schneid C. maximowiczii Schneid C. sanguinea Pall. C. dahurica Schneid C. laevigata Poir. M. germanica L. Laevigata Mespilus C.scabrifolia Rehd. C.jozana. Schneid
Plotype
No. of Species
No. of Sample
ID
Taxon
Region
Group
H1
3
7
HGLR/FLH/S4/MSZ/NA
C. chlorosarca Maxim.
NC
Sanguineae
H2
1
3
AET
C. altaica (Loud.) Lange.
NC
Sanguineae
H3
3
6
GSSZ/GYSZ/LNSZ
C. kansuensis Wils.
NC
Sanguineae
H4
2
2
ZWSLH/GSSZ
ZWSLH/ GSSZ
NC
Sanguineae
H5
1
1
J6
J6
NA
H6
1
2
HSD
HSD
NA
H7
3
3
DZSZ/HH/germanica
C. monogyna Jacq.
EU
H8
1
1
ZGE
C. songarica Koch
NC
Orientales
H9
1
1
JT
C. hupehesis Sarg.
SC
Henryanae
H10
2
8
HBSZ/YNSZ
C. hupehesis Sarg.
SC
Henryanae
H11
1
4
AG
C. cuneata Sieb.et Zucc
SC
Henryanae
H12
1
12
ZD/ZS/YSZ
C. cuneata Sieb.et Zucc
SC
Henryanae
H13
1
3
XBRZ/SHSLH/JSTSLH
C. pinnatifida Bunge
Wide
Pinnatifidae
H14
1
2
SZ/HGSLH
C. pinnatifida Bunge
Wide
Pinnatifidae
H15
2
53
PIN/MHSLH
C. pinnatifida Bge. Major N.E.Br.
Wide
Pinnatifidae
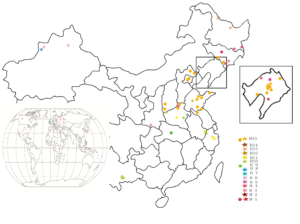
Figure 4: Chloroplast haplotype distribution map of 108 Crataegus.
Analysis of Chloroplast Gene Fragment Sequence Variation Sites
Through sequence analysis and mutation site analysis, a total of 23 mutation sites were detected, including 16 point mutations and 7 insertions / deletions between different species. In the mode of point mutation, there are 4 point mutations in trnG-trnS, including 3 transversions and 1 transformation; all 4 point mutations in psbA-trnH were transversions; there are 8 point mutations in trnH-trnK, including 3 transversions and 5 conversions.
Among the insertions and deletions among different species, C. maximowiczii has a unique sequence ATTTGTTTTATTTGTTTT at 160bp-177bp in psbA-trnH region, and all or part of the fragments are missing in other resources; C. brettschneideri has the unique sequence TTCTTTATTCCTTTTATTTTA at 304bp-324bp in trnG-trnS region, which is not found in other Hawthorn resources; C. monogyna has the unique sequence CAATAAATATAGATA-TTCAATAATT at 59bp-82bp in trnH-trnK region, which is not found in other Hawthorn resources. The sequence CAATAAATATAGATATT at 83bp-98bp in trnH-trnK position
Nucleotide
psbA-trnH
trnG-trnS
trnH-trnK
107
126
129
160-177
194
7
211
304
341-369
456
477
649
7
53
59
83
136
223
625
763
814
817
H1
G
T
G
●ATTTGTTTTATTTGTTTT
T
G
–
▲
-AAATAACG
G
A
C
G
TC
–
–
G
T
A
A
C
A
H2
.
.
.
——————
.
.
.
–
.
T
.
.
.
.
.
□
.
.
.
.
.
.
H3
.
.
.
ATTT———GTTTT
.
.
.
–
A——GTA
.
.
.
.
.
.
.
A
.
.
.
.
.
H4
.
.
.
——————
.
.
◎
–
.
.
.
.
.
.
.
□
.
.
.
.
.
.
H5
.
A
.
——————
A
.
.
–
A——GTA
.
.
.
.
.
.
□
.
A
.
.
T
.
H6
.
.
T
ATTT———GTTTT
.
.
.
–
.
.
.
.
.
.
.
□
.
A
.
.
.
.
H7
T
.
.
ATTTGTTTT—TGTTTT
.
.
.
–
A——GTA
.
.
T
–
.
■
□
.
A
.
.
.
G
H8
T
.
.
ATTTGTTTT—TGTTTT
.
.
.
–
A——GTA
.
.
T
–
.
.
□
.
A
.
.
.
G
H9
.
.
.
ATTTGTTTT—TGTTTT
.
.
.
–
.
.
.
.
.
TA
.
□
.
A
.
C
.
.
H10
.
.
.
ATTTGTTTT—TGTTTT
.
C
.
–
A——GTA
.
.
.
.
TA
.
□
.
A
.
C
.
.
H11
.
.
.
ATTTGTTTT—TGTTTT
.
C
.
–
A——GTA
.
.
.
.
.
.
□
.
A
.
.
.
.
H12
.
.
.
ATTTGTTTT—TGTTTT
.
C
.
–
A——GTA
.
.
.
.
.
.
□
.
A
.
.
.
.
H13
.
.
T
ATTTGTTTT—TGTTTT
.
C
.
–
.
.
.
.
.
GC
.
□
.
A
G
.
.
.
H14
.
.
T
ATTTGTTTT—TGTTTT
.
C
.
–
A——GTA
.
.
.
.
GC
.
□
.
A
.
.
.
.
H15
.
.
T
ATTTGTTTT—TGTTTT
.
C
.
–
A——GTA
.
C
.
.
GC
.
□
.
A
.
.
.
.
(Notes:. represents the same base as H1, and the replaced base or base sequence is marked in different places; ● represents the sequence of C.maximowiczii only found in H1, and the sequence in this region of C.brettschneideri is the same as that in H3; ▲ represents the unique sequence (TTCTTTATTCCTTTTATTTTA) in C. brettschneideri, and other species do not have this sequence; ◎ represents the unique sequence (GATTTCTATCTTTA) in zwslh and gssz in H4; ■ represents the unique sequence (CAATAAATATAGATATCAATAATT of C. monogyna in H7, and there is no such sequence in other species; □ represents the sequence (CAATAAATATAGATAT))
Region has not in H1 and H3 haplotypes, and other tested materials contain this sequence fragment. The statistical information of variation sites in the three chloroplast gene spacer regions is shown in table 5.
Analysis of the Haplotype Network Graph
The haplotype network graph (Figure 5 and Figure 6) analysis showed that, in the origin of chloroplasts, hawthorn samples from northern China with haploids H1–H4 had the same ancestral haplotype (no samples were collected in the present study). Hawthorn samples from northern China with haploids H9–H15 had the same ancestral haplotype of H12. Therefore, the haplotypes of hawthorn from northern and southern China were different, indicating that the northern and southern China Crataegus had different origins. The European haplotype H7, having an earlier divergence time, constituted a sister group with southern China Crataegus; American haplotypes H5 and H6, with earlier divergence time, constituted one with northern China Crataegus.
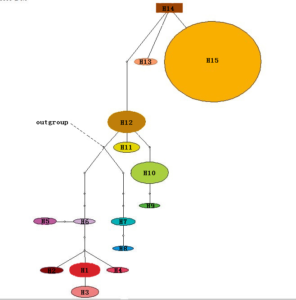
Figure 5: Chloroplast haplotype TCS network (Dots indicate cultivated hawthorn. Five stars means wild hawthorn).
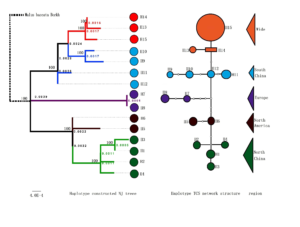
Figure 6: Chloroplast haplotype phylogenetic tree, haplotype network and region.
Notes:
H1-H15 indicates the Chloroplast haplotype contained in the tested 108 Crataegus.
Left : Chloroplast haplotype phylogenetic tree from neighbor joining with MEGA. Nodes (above branches) with bootstrap values >50% are indicated; nodes (right) are divergence times.
Middle : Chloroplast haplotype TCS network
Right : Sample distribution region
Discussion
Speciation and Origin of Hawthorn in China
Our study revealed that hawthorn species in northern and southern China formed two distinct branches. North American hawthorns were more closely related to those from northern China, whereas European hawthorns were more closely related to those from southern China. Furthermore, in chloroplast haplotype network diagram, the European haplotype (H7) was closer to the haplotype seen in southern China, and the haplotypes of species from North America (H5 and H6) were closer to the haplotype of those from northern China. And haplotypes associated with different regions were isolated from each other, and there was no interflow and sharing among haplotypes. Therefore, hawthorn in northern and southern China underwent different speciation events. The first occurred in northern China and the second occurred in southern China. The results of the present study were consistent with those of Du et al. [25]. The Crataegus taxa of southwestern species shared a gene pool with European Crataegus, and the northeastern species shared a gene pool with the North American species, suggesting that the tested specimens may have experienced two different speciation events. Wang [34] found the same southwestern route based on the plants from other genus and families. European haplotype H7 and American haplotypes H5 and H6 had an earlier divergence time (Figure 5), indicating that Crataegus might have originated in Europe and North America and then migrated from Europe into southern China and from North America into northern China by trans-Beringian migration. The occurrence of these two events might be related to the uplift and barrier of the Qinghai-Tibet Plateau, Hengduan Mountains, and others.
Molecular Relationship and Hybrid Parents of C. brettschneideri
Several studies have shown that C. brettschneideri was a rare triploid species [35] of Chinese hawthorn. Whether C. brettschneideri is a separate species or not has always been a controversial topic. Some researchers thought it was a variant of C. pinnatifida relaying on the evidence obtained in physiological and molecular studies. For example, Guo et al. [36] suggested that C. brettschneideri was more closely related to C. pinnatifida based on peroxidase is ozymograms. Dai et al. [27] also believed that C. brettschneideri was a variant of C. pinnatifida based on Random Amplified Polymorphism DNA analyses. Other studies suggested it should be considered a separate species; Wu [37] concluded that C. brettschneideri was an independent species based on the results of cp-RFLP. Du et al. [25] hypothesized that C. brettschneideri might be a new species due to hybridization analyses between C. pinnatifida and C. maximowiczii, based on geography (i.e., C. brettschneideri is distributed at the border of C. maximowiczii and C. pinnatifida), tree mix genetic analysis (which indicated that gene flow had occurred from C. maximowiczii to C. brettschneideri), and STRUCTURE analysis (which indicated that C. brettschneideri shared a gene pool with C. pinnatifida and C. maximowiczii).
In the present study, the cp-ITS combined molecular evolutionary tree showed that C. brettschneideri separated from C. pinnatifida and C. maximowiczii, and formed a separate branch, and thus C. brettschneideriformed a separate species condition. Further, separate combined chloroplast gene molecular analysis showed that C. brettschneideri and C. maximowiczii were closely related and belonged to the same haplotype, H1. Separate ITS molecular analysis showed that C. brettschneideri and C. pinnatifida had a close genetic relationship and shared the haplotype H15. Chloroplast genes contain the feature of maternal inheritance, yet ITS genes show more nuclear characteristics of the hybrid; also, C. maximowiczii was a tetraploid species [38] and C. pinnatifida was a diploid species [35, 38-41], and both were often distributed together. Therefore, we speculate that C. maximowiczii was the female parent of C. brettschneideri and C. pinnatifida was the male parent of C. brettschneideri. Thus, we believe that C. brettschneideri is not a C. pinnatifida variety, but rather an independent new species of natural hybridization, with C. pinnatifida as the male parent and C. maximowiczii as the female parent.
Preliminary Research on the Classification Status of Crataegus ZWSLH and GSSZ
The two endemic taxa of ZWSLH and GSSZ have been controversial in the classification system of hawthorns because of their particularity. In the present study, the results of associative molecular analysis based on chloroplast and ITS gene fragments were different from those based on the morphological feature classification system. Combined molecular analysis based on chloroplast gene fragments showed that ZWSLH and GSSZ were closely related and clustered as a single branch of the Sanguineae group and were closely related to C. maximowiczii, consistent with the findings of Du et al. [25]. The two taxa showed higher similarity to C. maximowiczii and C. sanguinea in the nSSR dendrogram and the two taxa clustered into a branch of northern China hawthorn species. The analysis of the chloroplast haplotype showed that they had the unique haplotype H4. Based on these results, we speculate that both ZWSLH and GSSZ had the bloodline of northern China hawthorn species and the same or more recently evolved species in chloroplast origin, and they belonged to the Sanguinea group with close ties of consanguinity to C. maximowiczii.
The analyses of ITS gene fragments showed that ZWSLH and GSSZ constituted different groups. GSSZ was still a part of the branch with the northern China hawthorn and was closely related to C. maximowiczii, whereas ZWSLH was linked to C. monogyna from Russia, C. laevigata from Britain, and M. germanica from the Czech Republic; therefore, ZWSLH had a European descent. If they were hybrid offspring, we speculate that they had different male ancestors. The male parent of GSSZ belonged to the genetic lineage of C. maximowiczii, whereas the male parent of ZWSLH was of European descent. Thus, we propose that GSSZ originated from the northern China hawthorn species and is closely related to C. maximowiczii. ZWSLH might be a hybrid of C. maximowiczii and the European hawthorn. These two groups had independent evolutionary branches and were proposed as two new germplasms.
Hypotheses from the Chloroplast Haplotype
It has been suggested that C. maximowiczii is as the section of the Sanguineae group [11]. However, the results of the present study did not support this idea, and the Sanguineae group might have another haplotype of origin. Philips [3] believed that C. scabrifolia evolved into most of the hawthorn species in China and Europe. The STRUCTURE analysis [25] showed that C. scabrifolia, the earliest species, belonged to the same lineage as the European species C. laevigata. However, our results showed that the chloroplast haplotype of C. scabrifolia and C. hupehensis belonged to the same one, H10, and they both originated from H12 (C. cuneata), which may be the ancestral haplotype of hawthorn in southern China.
The chloroplast haploid type network diagram showed that H14 was the older chloroplast haploid type, which was not consistent with C. pinnatifida being a relatively new evolutionary type. We thought that H14 might be the direct origin haploid of Pinnatifidae group based on the results of the present study, which related to the long cultural history of C. pinnatifida major.
Conclusion
Our analyses suggest Crataegus in northern and southern China have respective origin relations. C. pinnatifida is part of the Pinnatifidae group and most members of this group were widely distributed cultivated species; C. pinnatifida Bge. Var. Major originates from C. pinnatifida. Incongruence between chloroplast and nuclear data support the hypothesis of a hybrid origin for C. brettschneideri. C. brettschneideri could be a separate species of natural hybridization with C. pinnatifida as the male parent and C. maximowiczii as the female parent. C. hupehensis and C. scabrifolia belong to the Henryanae group and their chloroplast haplotypes are both H10, which originate from haplotype H12 (C. cuneata). C. cuneata has a molecular evolutionary origin earlier than that of C. scabrifolia. The findings of this study provide valuable information on the genetic relationship and chloroplast haplotype origin of Crataegus in China.
Acknowledgments
We thank Lyu Deguo, National Fruit-tree Germplasm Resourecs-Shenyang Hawthorn Repository of China, and Professor Fu, Professor Zhao, Dr. Liu Luxian, Zhejiang University for the support of assisting with the experiments. We thank Dr. Zhang Qijing, Dr. Qiu-ping Zhang, Liaoning Institute of Pomology, for analyzing data and commenting on the manuscript. And we also thank our editor for language, grammar, and improved clarity.
Funding: This work was supported by the Program (project) of “The conservation and utilization of crop germplasm resource-Hawthorn (2015-2017)”
References
- Dickinson TA, Proctor S, Shipley PR, Lee J, Coughlan J, et al. (2014) “Natural health products and Crataegus of the Pacific Northwest,” In Proceedings of the Annual Meeting, Canadian Botanical Association, Montreal, Qc.
- Edwards JE, Brown PN, Talent N, Dickinson TA, Shipley PR (2012) A review of the chemistry of the genus Crataegus. Phytochemistry 79: 5-26.
- Phipps JB (1990) Mespilus canescens, a new Rosaceous endemic from Arkansas. Syst. Bot. 15: 26-32.
- DeVore ML, Pigg KB (2007) A brief review of the fossil history of the family Rosaceae with a focus on the Eocene Okanogan Highlands of eastern Washington State,USA, and British Columbia, Canada. Plant Systematics and Evolution 266: 45-57.
- Zhao HC, Feng BT (1996) China fruit plant monograph of Hawthorn (Crataegus) flora. China Forestry Publishing House, Beijing.
- Xin X, Zhang Y M (1997) Chinese hawthorn germplasm resources and utilization. China Agricultural Press, Beijing.
- Dong WX, Li ZX (2015) The Science and Practice of Chinese Fruit Tree: Hawthorn. Shanxi: Science Press. pp 23-52.
- Phipps JB (1983) Biogeographic, taxonomic, and cladistic relationships between East Asiatic and North American Crataegus. Ann. Mo. Bot. Gard. 70: 667-700.
- Evans RC, Campbell CS (2002) The origin of the apple subfamily (Maloidea; Rosaceae) is clarified by DNA sequence data from duplicated GBSSI genes. Am. J. Bot. 89: 1478-1484.
- Lo EYY, Sasa S, Christensen K (2009) Evidence for genetic association between East Asian and western North American Crataegus (Rosaceae) and rapid divergence of the eastern North American lineage based on multiple DNA sequences. Molecular Phylogenetics and Evolution 51: 157-168.
- Zarrei M, Talent N, Kuzmina M, Lee J, Lund J, et al. (2015) DNA barcodes from four loci provide poor resolution of taxonomic groups in the genus Crataegus. AoB PLANTS.
- Güney M, Kafkas S, Keles H, Aras S, Ercişli S (2018) Characterization of hawthorn (Crataegus spp.) genotypes by SSR markers. Physiology and Molecular Biology of Plants 24: 1221-1230.
- Khiari S, Boussaid M, Messaoud C (2015) Genetic diversity and population structure in natural populations of Tunisian Azarole (Crataegusazarolus, L. var. aronia L.) assessed by microsatellite markers. Biochem. Syst. Ecol 59: 264-270.
- Brown JA, Beatty GE, Finlay C, Montgomery I, Tosh DG, et al. (2016) Genetic analyses reveal high levels of seed and pollen flow in hawthorn (Crataegus monogyna, Jacq.), a key component of hedgerows. Tree Genetics & Genomes 12: 58.
- Erfani-Moghadam J, Mozafari M, Fazeli A (2016) Genetic variation of some hawthorn species based on phenotypic characteristics and RAPD marker. Biotechnol. Biotech. Equip 30: 247-253.
- ZArei A, Erfanimoghadam J, Mozaffari M (2017) Phylogenetic analysis among some pome fruit trees of Rosaceae family using RAPD markers. Biotechnology and Biotechnological Equipment.
- Sheng F, Chen SY, Tian J, Li P, Qin X, et al. (2017) Morphological and ISSR molecular markers reveal genetic diversity of wild hawthorns (Crataegus songorica Koch.) in xinjiang, China. J. Inte. Agri. 10: 2402-2406.
- Emami A, Shabanian N, Rahmani MS, Khadivi A, Mohammad-Panah N (2018) Genetic characterization of the Crataegus genus: Implications for insitu conservation. Scientia Horticulturae 231: 56-65.
- Wu FF, Zhang ZH, Dai HY, Zhang Y, Zhang LL (2008) Genetic relationship of some Crataegus (Hawthorn) revealed by chloroplast DNA PCR-RFLP. Journal of Biotechnology.
- Zhang Y, Dai HY, Zhang QJ, Li H, Zhang ZH (2008) Assessment of genetic relationship in Crataegus genus by the apple SSR primers. J. Fruit. Sci 25:521-525.
- Julia Ferm (2019) A preliminary phylogeny of Zapoteca (Fabaceae: Caesalpinioideae:Mimosoid clade). Plant Systematics and Evolution 305: 341-352.
- Li L, Shih‑Wen C, Li B, Song‑Jun Z, Hai‑Fei Y, Shi‑Jin L (2020) New insight into the molecular phylogeny of the genus Liparis s.l. (Orchidaceae: Malaxideae) with a new generic segregate: Plant Systematics and Evolution 306: 54.
- Su K, Guo YS, Wang G, Zhao YH, Dong WX (2015) Genetic diversity analysis of fruit characteristics of hawthorn germplasm. Genet. Mol. Res. 14: 16012-16017.
- Ma SLY, Lu YM (2016) Classification and phylogenetic analysis of Chinese hawthorn assessed by plant and pollen morphology. Gen.Mol.Res.
- Du X, Zhang X, Bu HD, Zhang TC, Lao Y C, et al. (2019) Molecular analysis of evolution and origins of cultivated hawthorn (Crataegus) and related species in China. Frontiers in Plant Science. 10: 443.
- Kyoko Yamane, Taihachi Kawahara (2018) Size homoplasy and mutational behavior of chloroplast simple sequence repeats (cpSSRs) inferred from intra- and interspecific variations in four chloroplast regions of diploid and polyploid Triticum and Aegilops Genetic Resources and Crop Evolution 65: 727-743.
- Dai HY (2007) Molecular Identification and Enhancement of Germplasms in Hawthorn. Ph.D. Thesis, Shenyang Agricultural University, Shenyang.
- Gu C, Spongberg SA (2003) Crataegus Linnaeus. In: Wu ZY, Raven PH, Hong DY (Eds) Flora of China Vol 9 (Pittosporaceae through Connaraceae). Science Press, Beijing and Missouri Botanical Garden Press, St. Louis, 111-117.
- Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13-15.
- Demesure B, Sodzi N, Petit R J (1995) A set of universal primers for amplification of polymorphic noncoding regions of mitoehondrial and ehloropllstDNA. Plants Molecular Ecology 4: 129-131.
- Müller K (2005) Incorporating information from length-mutational events into phylogenetic analysis. Mol. Phylogenet. Evol 38: 667-676.
- Swofford DL (2002) PAUP4.0B10: Phylogenetic Analysis Using Parsimony Sunderland Sinauer Associates. MA.
- Pickrell JK, Pritchard JK (2012) Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet.
- Wang WT (1992) On some distribution patterns and some migration routes found in the eastern Asiatic region. Act Phy Sin. 30: 1-24.
- Xin X G (1991) Study on the chromosome number of Crataegus and cultivars in China. Journal of Shenyang Agricultural University 22: 27-35.
- Guo TJ, Jiao PJ (1995) Hawthorn (Crataegus) resources in China. Hortscience 30: 1132-1134.
- Wu FF (2009) Genetic diversity of Crataegus revealed by chloroplast DNA. M.S. Thesis, Shenyang Agricultural University, Shenyang.
- Pu FS, Lin SH, ZHang DX (1987) Observation on chromosome number of some species and varieties of Crataegus in China. Chins Fruits.
- Song WQ, Li XL, CHen RY, Lin SH, ZHang DX, Pu FS (1985) Study on chromosome number of some hawthorn plants in China. Acta Horticulturae Sinica 2:73-76.
- Hamilton MB (1999) Four primers pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Mol Ecol Notes 8:513-525.
- Zhang QP, Wei X, et al. (2018) The genetic relationship and structure of some natural interspecific hybrids in Prunus subgenus Prunophora, based on nuclear and chloroplast simple sequence repeats. Genetic Resources and Crop Evolution 65: 625-636.